Memory management in Python#
Andrew Delman, updated 2024-07-27
Objectives#
Demonstrate how Python objects (variables) are stored in and cleared from memory, and establish good practices for conserving memory in a Python workspace.
Introduction#
Python is a very useful computing language for doing numerical analysis of large datasets, largely because of packages that have been developed specifically for this purpose. Xarray allows us to open data from a number of files using a single line of code (with open_mfdataset), while Dask makes it easy to parallelize computations and stagger memory usage by “chunking” the computations. However, we still need to be mindful of the memory limitations of our workspace, and there are some quirks to memory management in Python.
Datasets to download#
If you don’t have them already, you may need to download the following datasets for the last part of this tutorial.
ECCO_L4_GEOMETRY_LLC0090GRID_V4R4
ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4 (1992-2017)
View vs. copy#
In some numerical computing languages such as Matlab, assigning a new variable instantly creates an independent block of memory in our workspace associated with that variable. In Python, when you create a variable by assigning data from another variable, the assignment does not create a copy of that data. Rather, a pointer called a view is created from the new variable to the data in the original. This is the default behavior for native Python objects such as lists, tuples, and dictionaries, as well as NumPy arrays.
Native Python object (list)#
Consider a simple list of 3 numbers
test_list = [1,2,3]
view_list = test_list
view_list
[1, 2, 3]
We can see if view_list is read from the same object in memory as test_list, using the is operator.
view_list is test_list
True
Not only that, because view_list is identical in memory space to test_list, when view_list is modified test_list is modified as well.
view_list[1] = 4
test_list
[1, 4, 3]
Compare this with creating a copy of test_list. There is a copy package that does this in Python, but you can also create a copy just by appending .copy(), or by using an operator on the right-hand side of the assignment. For example, we can “append” an empty list to create a copy.
test_list = [1,2,3]
copy_list = test_list + []
copy_list
[1, 2, 3]
Note that if we use the == operator, which compares the values of the two objects, the result is True.
copy_list == test_list
True
But if we compare using the is operator, we find that the two objects are independent.
copy_list is test_list
False
And so if copy_list is modified, test_list is not changed.
copy_list[1] = 4
test_list
[1, 2, 3]
NumPy arrays#
Views or copies of NumPy arrays can be created using the same general syntax used above.
import numpy as np
test_array = np.array([1,2,3])
view_array = test_array
view_array
array([1, 2, 3])
view_array is test_array
True
view_array[1] = 4
test_array
array([1, 4, 3])
test_array = np.array([1,2,3])
copy_array = test_array + 0
copy_array
array([1, 2, 3])
copy_array == test_array
array([ True, True, True])
copy_array is test_array
False
copy_array[1] = 4
test_array
array([1, 2, 3])
For NumPy arrays, views or copies can also be created by appending .view() or .copy() to the variable name. For more examples of NumPy array views/copies and how they are handled by Xarray, see the previous tutorial.
Memory footprint#
As the examples above showed, understanding the difference between a view and a copy is important to ensure your code is working the way you think it should be. Python’s capacity to reference an object without creating a separate copy of it is also a helpful memory-saver. But there are other ways to create a Python object without writing its data to memory. Let’s consider the memory footprint of two types of arrays: NumPy arrays and Dask arrays.
Note: There are a number of tools that can be used to estimate the memory footprint of an object in Python, such as the object’s
__sizeof__method,sys.getsizeof, andpympler.asizeof. To get the most accurate estimate of actual memory usage in the workspace, in this tutorial we usepsutil.virtual_memoryfrom the [psutil]((https://psutil.readthedocs.io/en/latest/#memory) package. This function tells us the memory available, and then we can track how it changes.
import numpy as np
import xarray as xr
import psutil
import os
# memory stats (in bytes)
psutil.virtual_memory()
svmem(total=7952175104, available=6066610176, percent=23.7, used=1612763136, free=2748981248, active=3048009728, inactive=1532313600, buffers=1011712, cached=3589419008, shared=8822784, slab=129548288)
The above output gives us a variety of information about the memory in our system. We will be tracking the available memory (given as bytes in the tuple above).
NumPy arrays#
Consider the view/copy distinction in the previous section, but with larger arrays.
# function to log memory
# and print change in available memory since last log point
def memory_log_print_avail_change(memory_log,\
print_before_avail='Change in available memory:'):
memory_log.append(psutil.virtual_memory())
print(print_before_avail,np.diff(np.asarray([memory_log[ind].available\
for ind in range(-2,0)]))[0]/(10**6),' MB')
memory_log = [psutil.virtual_memory()]
test_array = np.reshape(np.arange(0,1000000),(1000,1000)).astype('float64')
memory_log_print_avail_change(memory_log)
Change in available memory: -8.155136 MB
The change in our available memory is about 8 MB, which is what we would expect for an array with \(10^6\) entries of type float64 (8 bytes each). How does our available memory change if we create a view of this array?
view_array = test_array
memory_log_print_avail_change(memory_log)
Change in available memory: -0.012288 MB
Effectively zero change! Note that we wouldn’t expect the change to be identically zero, since a pointer has been created somewhere in memory. But the actual change in available memory depends not only on the memory footprint of new object(s) but on how Python allocates memory in response to those changes.
Now what is the memory impact (footprint) from making a copy of this array?
copy_array = test_array.copy()
memory_log_print_avail_change(memory_log)
Change in available memory: -7.725056 MB
Very close to the size of the original array.
Dask arrays and delayed computations#
You may have noticed that NumPy creates (non-memory consuming) views of an array only if:
the view references the original array without any operations applied to it, OR
the view references a subset of the original array using simple indexing
So what if you want to create a “view” of an array that has been operated on in any variety of ways, while minimizing the memory impact until you are ready to actually “execute” the computations? Enter Dask arrays. Let’s create a Dask array that is a view of our test_array, and then operate on it (take the square root).
import dask.array as da
memory_log.append(psutil.virtual_memory())
dask_test_array = da.from_array(test_array,chunks=(100,100)) # array has chunks of size 100x100
dask_sqrt_array = dask_test_array**(1/2)
memory_log_print_avail_change(memory_log)
Change in available memory: 0.0 MB
No new memory usage from the two operations above. And now, we compute dask_sqrt_array so that it becomes a NumPy array in memory:
sqrt_array_computed = dask_sqrt_array.compute()
memory_log_print_avail_change(memory_log)
Change in available memory: -26.894336 MB
Notice that the memory usage from calling .compute is larger than we would expect for the size of the array: (1000 x 1000 elements) x (8 bytes/element) \(\approx\) 8 MB. But there is some overhead associated with calling .compute for the first time in a notebook or Python script/function. What if we perform the same computation as above, using copy_array as our source since it is independent in memory?
dask_copy_array = da.from_array(copy_array,chunks=(100,100)) # array has chunks of size 100x100
dask_copy_sqrt_array = dask_copy_array**(1/2)
copy_sqrt_computed = dask_copy_sqrt_array.compute()
memory_log_print_avail_change(memory_log)
Change in available memory: -8.8064 MB
Much closer to our expected memory usage, since it is not the first time .compute has been called on a Dask array in this notebook.
Let’s try a “reduction” operation, where the computed result is smaller than the intermediate arrays involved. In this case we will take the square of dask_test_array and then sum over one of the dimensions.
dask_sq_array = dask_test_array**2
sq_sum_computed = (dask_sq_array.sum(axis=-1)).compute()
memory_log_print_avail_change(memory_log)
Change in available memory: -2.105344 MB
The memory usage is not as small as the expected size of sq_sum_computed (0.008 MB), but it is much smaller than the 8 MB that each of the intermediate arrays would have used if they were regular NumPy arrays. Calling .compute on a reduced array like this is where Dask really helps us save memory.
Deleting variables to free up memory#
Now let’s say we want (or need) to increase our available memory by deleting some of the arrays that we just computed. In Matlab for example you might just call clear variable to free up the memory used by variable. But in Python it is not that simple, especially when Dask is involved.
NumPy case (no Dask)#
Here we will compute (first using NumPy only) and then delete the square root of test_array, and compare the memory impacts to when we get Dask involved.
# restart the memory log
memory_log = [psutil.virtual_memory()]
# copy test_array (to create a fresh "source" for this calculation)
test_numpy = test_array.copy()
memory_log_print_avail_change(memory_log)
# take the square root
test_numpy_sqrt = test_numpy**(1/2)
memory_log_print_avail_change(memory_log)
# delete the new square root array
del test_numpy_sqrt
memory_log_print_avail_change(memory_log)
Change in available memory: -7.897088 MB
Change in available memory: -8.00768 MB
Change in available memory: 0.0 MB
We created two new NumPy arrays in memory, and the footprint of each is ~8 MB, what we would expect. But when we deleted one of these arrays, we didn’t recover its memory. Why?
To understand this, let’s consider how Python does “garbage collection”. Each Python object has a certain number of references, which are created by equations/operational relationships to other objects). del decreases an object’s reference count by 1, but the garbage collector will only free up memory associated with an object if the object and all objects it is associated with are zero.
We can use sys.getrefcount to determine an object’s number of references, but calling sys.getrefcount on an object also creates a temporary reference to that object. So the “true” number of references will be 1 less than the output of sys.getrefcount.
# Recover test_numpy_sqrt (since we didn't really delete it, it is still cached in memory)
test_numpy_sqrt = test_numpy**(1/2)
memory_log_print_avail_change(memory_log)
import sys
print('Reference count of test_numpy_sqrt:',sys.getrefcount(test_numpy)-1)
print('Reference count of test_numpy:',sys.getrefcount(test_numpy)-1)
Change in available memory: 0.0 MB
Reference count of test_numpy_sqrt: 1
Reference count of test_numpy: 1
These arrays each have 1 reference, which means if we use del on both of them, and they are not referencing any other object, then we should be able to clear both of them from memory.
del test_numpy_sqrt
del test_numpy
memory_log_print_avail_change(memory_log)
Change in available memory: 17.256448 MB
And indeed, we did recover almost 16 MB of memory, the equivalent of the 2 arrays.
Complications of Dask#
We just demonstrated that in NumPy, when you execute a line of code that involves multiple objects, you usually establish a reference between them so that to delete the memory content of one, you have to delete all of them. With Dask, it gets still more complicated. Let’s try to delete copy_sqrt_computed, which has the same values as test_numpy_sqrt did but was computed using Dask arrays.
print('Reference count of copy_sqrt_computed:',sys.getrefcount(copy_sqrt_computed)-1)
print('Reference count of dask_copy_sqrt_array:',sys.getrefcount(dask_copy_sqrt_array)-1)
print('Reference count of dask_copy_array:',sys.getrefcount(dask_copy_array)-1)
print('Reference count of copy_array:',sys.getrefcount(copy_array)-1)
Reference count of copy_sqrt_computed: 1
Reference count of dask_copy_sqrt_array: 1
Reference count of dask_copy_array: 1
Reference count of copy_array: 102
All of the above arrays are linked by a chain of references, and notice that copy_array has 102 references! How did that happen? The Dask array dask_copy_array consists of 10x10 = 100 chunks, each sourced from copy_array. Each of those chunks adds a reference to copy_array.
Let’s delete all of the above arrays, as we did in the previous case with NumPy. As we do this, we will also track the number of references to copy_array.
del copy_sqrt_computed
del dask_copy_sqrt_array
del dask_copy_array
print('Reference count of copy_array: ',sys.getrefcount(copy_array)-1)
del copy_array
memory_log_print_avail_change(memory_log)
Reference count of copy_array: 1
Change in available memory: 0.0 MB
Now we deleted what would seem to be all the arrays that could be linked by references to copy_sqrt_computed, and still we haven’t recovered memory!
Freeing up memory by saving to disk#
Python’s memory management can be difficult to understand sometimes, especially when sophisticated packages like Dask are involved. But one way that an array’s memory can be made available again is to save it to disk storage. Even if we don’t need the array’s data anymore, saving a NumPy array to a .npz archive moves the array’s reference away from the Dask array in the local workspace, to the newly created file.
Let’s try repeating the previous experiment by re-creating copy_array and make sure that new memory is being allocated for it. Don’t ask me why we need to call .copy 3 times before new memory is allocated for the array; maybe you know the answer, but I don’t!
copy_array = test_array.copy()
memory_log_print_avail_change(memory_log)
Change in available memory: 0.0 MB
copy_array_2 = copy_array.copy()
memory_log_print_avail_change(memory_log)
Change in available memory: 0.0 MB
copy_array_3 = copy_array_2.copy()
memory_log_print_avail_change(memory_log)
Change in available memory: -7.839744 MB
# re-create the in-memory array copy_sqrt_computed
dask_copy_array = da.from_array(copy_array_3,chunks=(100,100)) # array has chunks of size 100x100
dask_copy_sqrt_array = dask_copy_array**(1/2)
copy_sqrt_computed = dask_copy_sqrt_array.compute()
memory_log_print_avail_change(memory_log)
Change in available memory: -8.105984 MB
Now that we finally re-created copy_sqrt_computed, let’s remove its memory from our workspace by first saving it to .npz.
# save copy_sqrt_computed to a .npz archive
np.savez('copy_sqrt_array.npz',copy_sqrt_array=copy_sqrt_computed)
memory_log_print_avail_change(memory_log,\
'Change in available memory from saving copy_sqrt_computed to disk:')
# delete the file that was just created (assuming it isn't needed later)
os.remove('copy_sqrt_array.npz')
# delete reference for copy_sqrt_computed
del copy_sqrt_computed
memory_log_print_avail_change(memory_log,\
'Change in available memory from deleting copy_sqrt_computed:')
Change in available memory from saving copy_sqrt_computed to disk: -8.052736 MB
Change in available memory from deleting copy_sqrt_computed: 15.716352 MB
We recovered (most of) the memory associated with copy_sqrt_computed, though it required saving the array to disk first (which initially used more memory), and then deleting its reference. Does this procedure work with the other in-memory array copy_array_3?
# # try the same thing with copy_array_3
# save NumPy array to a .npz archive
np.savez('copy_array.npz',copy_array=copy_array_3)
memory_log_print_avail_change(memory_log,\
'Change in available memory from saving copy_array_3 to disk:')
# delete the file that was just created (assuming it isn't needed later)
os.remove('copy_array.npz')
# delete references involving copy_array_3
del dask_copy_sqrt_array
del dask_copy_array
del copy_array_3
memory_log_print_avail_change(memory_log,\
'Change in available memory from deleting copy_array_3:')
Change in available memory from saving copy_array_3 to disk: -7.716864 MB
Change in available memory from deleting copy_array_3: 15.745024 MB
The same procedure did not help to free up the memory from copy_array_3, which was probably too interconnected with Dask since a Dask array was created from it. Usually this is not a problem for memory management since we are more likely to load Dask arrays from files, rather than creating them from NumPy arrays already in memory.
Let’s take a look at the memory log and compare the case that did not involve Dask with the one that did.
import matplotlib.pyplot as plt
fig,ax = plt.subplots(1,2,figsize=(12,7))
ax[0].plot((np.asarray([memory_log[ind].used for ind in range(0,6)])\
- memory_log[0].used)/(10**6),color='r',label='used')
ax[0].plot((np.asarray([memory_log[ind].available for ind in range(0,6)])\
- memory_log[0].available)/(10**6),color='b',label='available')
ax[0].set_ylim([-27,27])
ax[0].axhline(y=0,color='k',lw=0.5)
ax[0].set_xlabel('Memory log entry')
ax[0].set_ylabel('Memory change since start (MB)')
ax[0].legend()
ax[0].set_title('No Dask')
ax[1].plot((np.asarray([memory_log[ind].used for ind in range(8,15)])\
- memory_log[8].used)/(10**6),color='r',label='used')
ax[1].plot((np.asarray([memory_log[ind].available for ind in range(8,15)])\
- memory_log[8].available)/(10**6),color='b',label='available')
ax[1].set_ylim([-27,27])
ax[1].axhline(y=0,color='k',lw=0.5)
ax[1].set_xlabel('Memory log entry')
ax[1].set_ylabel('Memory change since start (MB)')
ax[1].legend()
ax[1].set_title('With Dask')
plt.show()
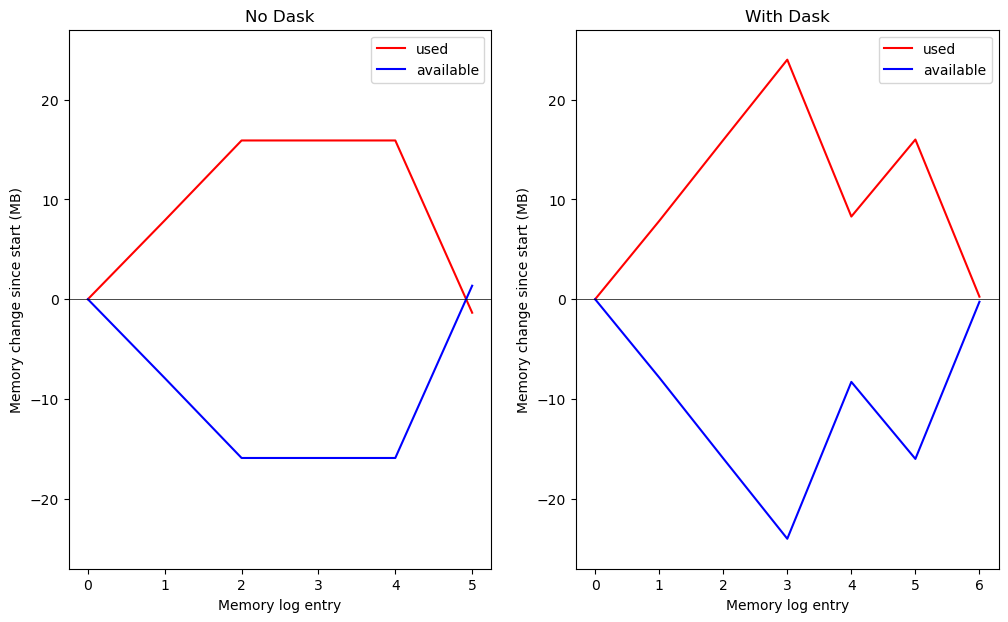
With NumPy only (no Dask) we were able to recover essentially all of the memory we used during the computation. With Dask we achieved a net loss of ~10 MB at entry #4 on the right (just over the size of one of the arrays) before losing more memory when saving copy_array_3 to disk.
It is worth remembering that Dask can also significantly help you limit the memory usage in your workspace (as we saw earlier with a reduction operation)…even though you relinquish some control over memory management when using it.
Xarray and memory management#
Unsolicited data caching#
The xarray package can open data files as Dask arrays using open_mfdataset, which is very useful for limiting memory usage in your workspace. xarray.open_mfdataset opens individual variables in a data file as Dask arrays, so that you get a “lazy” view of a file (or many files concatenated together) without immediately loading data into memory. However, when data chunks are loaded into memory using .compute, this data is typically cached to speed up future loads of the same data. This results in unintended memory usage that might be problematic depending on the size of the loaded data, the data chunks, and the limitations of your computing environment.
Consider a case where we load (compute) into memory the monthly sea surface temperature (SST) from ECCO, 1992-2017.
import numpy as np
import xarray as xr
from os.path import join,expanduser
import glob
import psutil
import ecco_access as ea
user_home_dir = expanduser('~')
# indicate mode of access
# options are:
# 'download': direct download from internet to your local machine
# 'download_ifspace': like download, but only proceeds
# if your machine have sufficient storage
# 's3_open': access datasets in-cloud from an AWS instance
# 's3_open_fsspec': use jsons generated with fsspec and
# kerchunk libraries to speed up in-cloud access
# 's3_get': direct download from S3 in-cloud to an AWS instance
# 's3_get_ifspace': like s3_get, but only proceeds if your instance
# has sufficient storage
access_mode = 's3_open_fsspec'
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to /efs_ecco/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join('/efs_ecco','ECCO_V4r4_PODAAC')
# for access_mode = 's3_open_fsspec', need to specify the root directory
# containing the jsons
jsons_root_dir = join('/efs_ecco','mzz-jsons')
ShortNames_list = ["ECCO_L4_GEOMETRY_LLC0090GRID_V4R4",\
"ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
# access datasets
StartDate = '1992-01'
EndDate = '2017-12'
ds_dict = ea.ecco_podaac_to_xrdataset(ShortNames_list,\
StartDate=StartDate,EndDate=EndDate,\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)
ecco_grid = ds_dict[ShortNames_list[0]]
ecco_TS = ds_dict[ShortNames_list[1]]
## merge grid and temperature datasets
ds = xr.merge((ecco_grid,ecco_TS[['THETA']]))
ds
<xarray.Dataset> Size: 7GB
Dimensions: (i: 90, i_g: 90, j: 90, j_g: 90, k: 50, k_u: 50, k_l: 50,
k_p1: 51, tile: 13, nb: 4, nv: 2, time: 312)
Coordinates: (12/21)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* k (k) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
* k_u (k_u) int32 200B 0 1 2 3 4 5 6 7 8 9 ... 41 42 43 44 45 46 47 48 49
... ...
Zu (k_u) float32 200B ...
Zl (k_l) float32 200B ...
XC_bnds (tile, j, i, nb) float32 2MB ...
YC_bnds (tile, j, i, nb) float32 2MB ...
Z_bnds (k, nv) float32 400B ...
* time (time) datetime64[ns] 2kB 1992-01-16T18:00:00 ... 2017-12-16T06:...
Dimensions without coordinates: nb, nv
Data variables: (12/22)
CS (tile, j, i) float32 421kB ...
SN (tile, j, i) float32 421kB ...
rA (tile, j, i) float32 421kB ...
dxG (tile, j_g, i) float32 421kB ...
dyG (tile, j, i_g) float32 421kB ...
Depth (tile, j, i) float32 421kB ...
... ...
hFacW (k, tile, j, i_g) float32 21MB ...
hFacS (k, tile, j_g, i) float32 21MB ...
maskC (k, tile, j, i) bool 5MB ...
maskW (k, tile, j, i_g) bool 5MB ...
maskS (k, tile, j_g, i) bool 5MB ...
THETA (time, k, tile, j, i) float32 7GB dask.array<chunksize=(1, 25, 7, 45, 45), meta=np.ndarray>
Attributes: (12/58)
acknowledgement: This research was carried out by the Jet...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-l...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute...
... ...
references: ECCO Consortium, Fukumori, I., Wang, O.,...
source: The ECCO V4r4 state estimate was produce...
standard_name_vocabulary: NetCDF Climate and Forecast (CF) Metadat...
summary: This dataset provides geometric paramete...
title: ECCO Geometry Parameters for the Lat-Lon...
uuid: 87ff7d24-86e5-11eb-9c5f-f8f21e2ee3e0# compute temperature for top level
memory_log = [psutil.virtual_memory()]
SST_computed = ds.THETA.isel(k=0).compute()
memory_log_print_avail_change(memory_log)
Change in available memory: -2155.773952 MB
But wait…should the temperature at 1 depth level take up ~2 GB of memory?
print('SST_computed data type:',SST_computed.dtype)
print('SST_computed number of elements:',SST_computed.size)
print('Estimated memory footprint of SST_computed:',(4*SST_computed.size)/(10**6),'MB')
SST_computed data type: float32
SST_computed number of elements: 32853600
Estimated memory footprint of SST_computed: 131.4144 MB
So why was so much more memory used than the size of the array itself? When .compute is invoked on a dataset sourced from file(s) using open_mfdataset, any chunk that is accessed is also cached (presumably to make subsequent loads of that data faster). So what are the chunk sizes in the k dimension?
k_dimnum = list(ds.THETA.dims).index('k')
ds.THETA.chunks[k_dimnum]
(25, 25)
The array is divided in 2 chunks in the k dimension each of size 25. So even if we are only using .compute on a single depth level, 25 depth levels are being cached (in a somewhat compressed format) and using up memory. Note that even if we re-chunk the dataset or data array after accessing the file(s), caching happens according to the native chunk sizes of the file. And at least in my experience, passing the option cache=False to open_mfdataset does not change the memory usage.
So what can we do? One option is to align our workflow more closely with the native chunking of the source file(s). At this point I recommend restarting the kernel of this notebook so we can get a fresh open of the data files.
%%time
import numpy as np
import xarray as xr
from os.path import join,expanduser
import glob
import psutil
import ecco_access as ea
user_home_dir = expanduser('~')
# indicate mode of access
# options are:
# 'download': direct download from internet to your local machine
# 'download_ifspace': like download, but only proceeds
# if your machine have sufficient storage
# 's3_open': access datasets in-cloud from an AWS instance
# 's3_open_fsspec': use jsons generated with fsspec and
# kerchunk libraries to speed up in-cloud access
# 's3_get': direct download from S3 in-cloud to an AWS instance
# 's3_get_ifspace': like s3_get, but only proceeds if your instance
# has sufficient storage
access_mode = 's3_open_fsspec'
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to /efs_ecco/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join('/efs_ecco','ECCO_V4r4_PODAAC')
# for access_mode = 's3_open_fsspec', need to specify the root directory
# containing the jsons
jsons_root_dir = join('/efs_ecco','mzz-jsons')
ShortNames_list = ["ECCO_L4_GEOMETRY_LLC0090GRID_V4R4",\
"ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
# access datasets
StartDate = '1992-01'
EndDate = '2017-12'
ds_dict = ea.ecco_podaac_to_xrdataset(ShortNames_list,\
StartDate=StartDate,EndDate=EndDate,\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)
ecco_grid = ds_dict[ShortNames_list[0]]
ecco_TS = ds_dict[ShortNames_list[1]]
## merge grid and temperature datasets
ds = xr.merge((ecco_grid,ecco_TS[['THETA']]))
# compute temperature for top level, by looping through time chunks
memory_log = [psutil.virtual_memory()]
SST_computed = ds.THETA.isel(k=0)
SST_computed_array = np.empty(SST_computed.shape).astype('float32')
for time_ind in range(ds.THETA.sizes['time']):
SST_computed_array[time_ind,...] = ds.THETA.isel(time=time_ind,k=0).compute()
SST_computed.data = SST_computed_array
memory_log.append(psutil.virtual_memory())
print('Change in available memory:',np.diff(np.asarray([memory_log[ind].available\
for ind in range(-2,0)]))[0]/(10**6),' MB')
Change in available memory: -1969.475584 MB
CPU times: user 40.3 s, sys: 4.21 s, total: 44.5 s
Wall time: 47.4 s
In the code above we did explicitly something that Dask does behind the scenes…looped through chunks so that our data loads are done in smaller pieces. But the change in memory is about the same as before, because by the end of the loop all of the top 25 depth levels of THETA have been cached.
A workaround to save workspace memory#
There is one more trick up our sleeves to try to flush out the cache…re-open the data files on each loop iteration using open_mfdataset. This should have the effect of replacing our cache-linked dataset object with a clean “lazy” dataset (no data loaded or cached).
And since calls to open_mfdataset involving hundreds of files take time, and this would be repeated hundreds of times, we will pickle the dataset object so that it can be re-used quickly. Essentially what we are doing is saving the “pointers” to the file contents, while flushing out the cache on each loop iteration.
Let’s restart the kernel one more time before running the following code.
%%time
import numpy as np
import xarray as xr
from os.path import join,expanduser
import glob
import psutil
import ecco_access as ea
user_home_dir = expanduser('~')
# indicate mode of access
# options are:
# 'download': direct download from internet to your local machine
# 'download_ifspace': like download, but only proceeds
# if your machine have sufficient storage
# 's3_open': access datasets in-cloud from an AWS instance
# 's3_open_fsspec': use jsons generated with fsspec and
# kerchunk libraries to speed up in-cloud access
# 's3_get': direct download from S3 in-cloud to an AWS instance
# 's3_get_ifspace': like s3_get, but only proceeds if your instance
# has sufficient storage
access_mode = 's3_open_fsspec'
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to /efs_ecco/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join('/efs_ecco','ECCO_V4r4_PODAAC')
# for access_mode = 's3_open_fsspec', need to specify the root directory
# containing the jsons
jsons_root_dir = join('/efs_ecco','mzz-jsons')
ShortNames_list = ["ECCO_L4_GEOMETRY_LLC0090GRID_V4R4",\
"ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
# access datasets
StartDate = '1992-01'
EndDate = '2017-12'
ds_dict = ea.ecco_podaac_to_xrdataset(ShortNames_list,\
StartDate=StartDate,EndDate=EndDate,\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)
ecco_grid = ds_dict[ShortNames_list[0]]
ecco_TS = ds_dict[ShortNames_list[1]]
# pickle ecco_TS dataset object
import pickle
ecco_TS_pickled = pickle.dumps(ecco_TS)
## merge grid and temperature datasets
ds = xr.merge((ecco_grid,ecco_TS[['THETA']]))
# compute temperature for top level, by looping through time chunks
# while re-opening and closing ecco_TS dataset on each loop iteration
memory_log = [psutil.virtual_memory()]
SST_computed = ds.THETA.isel(k=0)
SST_computed_array = np.empty(SST_computed.shape).astype('float32')
for time_ind in range(ds.THETA.sizes['time']):
# re-load pickled dataset object (equivalent to calling open_mfdataset again, but much quicker)
ecco_TS = pickle.loads(ecco_TS_pickled)
# compute SST for current time chunk
SST_computed_array[time_ind,...] = ds.THETA.isel(time=time_ind,k=0).compute()
# close dataset object
ecco_TS.close()
SST_computed.data = SST_computed_array
memory_log.append(psutil.virtual_memory())
print('Change in available memory:',np.diff(np.asarray([memory_log[ind].available\
for ind in range(-2,0)]))[0]/(10**6),' MB')
Change in available memory: -128.9216 MB
CPU times: user 1min 16s, sys: 4.84 s, total: 1min 21s
Wall time: 1min 28s
Our memory usage now is very close to the expected footprint of the SST array itself, rather than ~15 times larger! The trade-off is that it took a little longer to load the data into memory with the “opening” and closing of the dataset object on each iteration.
Note that it is ecco_TS that we have flushed the cache from, not ds. This procedure needs to be applied to the dataset object associated with the initial open_mfdataset call, rather than any subsequent datasets that it might be merged or concatenated into.
To review, the procedure to flush the cache associated with open_mfdataset is:
When loading data explicitly in smaller “chunks”, be sure to loop through the dataset in ways that are well aligned with the pre-existing chunking of the source files.
Before calling
.compute, “re-open” the dataset by callingopen_mfdataset(or a pickled equivalent).Load your chunk into memory using
.compute.Close the dataset object (this may not be strictly necessary, but is a good practice for clean coding).
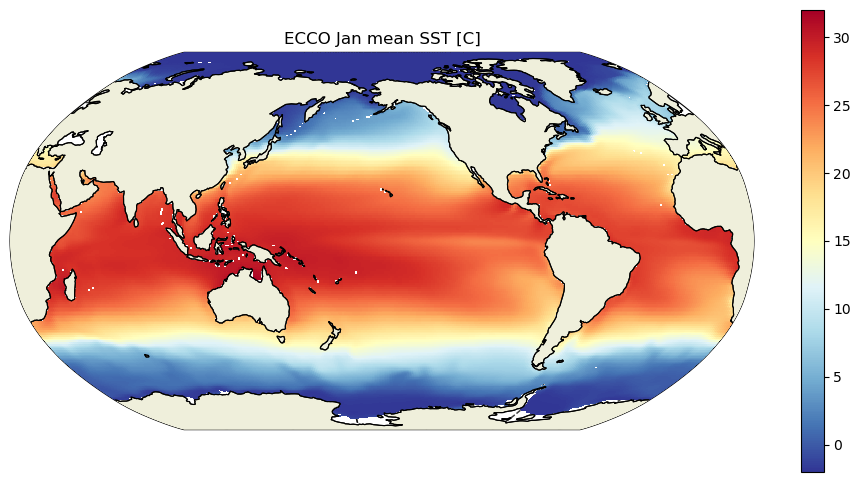
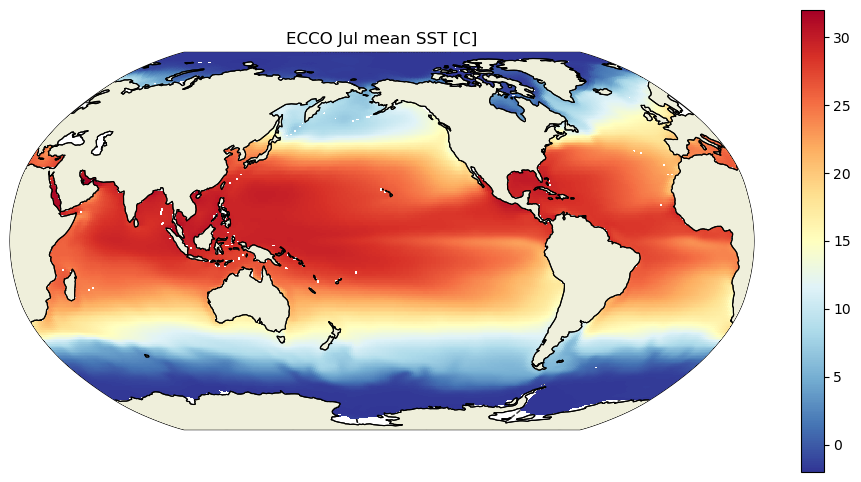
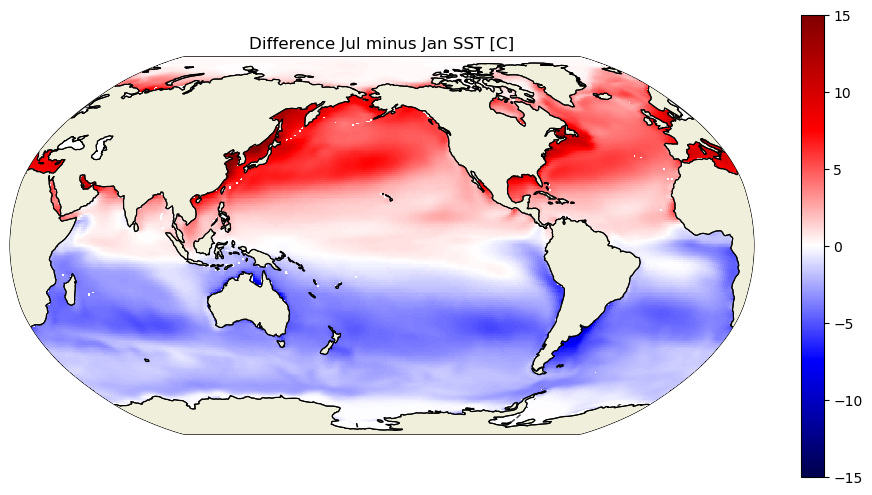
And now, just to be sure that it is SST that we have loaded, and because it is fun to end a tutorial by plotting data, let’s look at the ECCO climatological SST for January and July.
import matplotlib.pyplot as plt
import ecco_v4_py as ecco
dx = .25
dy = .25
plt.figure(figsize=(12,6))
ecco.plot_proj_to_latlon_grid(ds.XC,ds.YC,\
SST_computed.isel(time=slice(0,None,12)).mean('time'),\
dx=dx,dy=dy,user_lon_0=200,\
cmin=-2,cmax=32,show_colorbar=True,cmap='RdYlBu_r')
plt.title('ECCO Jan mean SST [C]')
plt.show()
plt.figure(figsize=(12,6))
ecco.plot_proj_to_latlon_grid(ds.XC,ds.YC,\
SST_computed.isel(time=slice(6,None,12)).mean('time'),\
dx=dx,dy=dy,user_lon_0=200,\
cmin=-2,cmax=32,show_colorbar=True,cmap='RdYlBu_r')
plt.title('ECCO Jul mean SST [C]')
plt.show()
plt.figure(figsize=(12,6))
ecco.plot_proj_to_latlon_grid(ds.XC,ds.YC,\
SST_computed.isel(time=slice(6,None,12)).mean('time')\
- SST_computed.isel(time=slice(0,None,12)).mean('time'),\
dx=dx,dy=dy,user_lon_0=200,\
cmin=-15,cmax=15,show_colorbar=True,cmap='seismic')
plt.title('Difference Jul minus Jan SST [C]')
plt.show()