Tutorial 3: Using the open-source Tapenade-generated adjoint of the MITgcm#
This is a brief description of using Tapenade to differentiate MITgcm. All experiments require a similar procedure, so without loss of generality, we describe the steps for the verification experiment tutorial_global_oce_biogeo running on a Linux platform. These steps are also outlined in the appendix of a recently published paper on MITgcm-AD v2 here.
Building the MITgcm#
There are multiple ways to build the MITgcm, as described in section 3.2 of the MITgcm documentation. We use git here to get the latest MITgcm tag, checkpoint68u.
$ git clone https://github.com/MITgcm/MITgcm.git
We then get the latest tagged release, \code{checkpoint68u} to ensure the reproducibility of results in the future.
$ git checkout checkpoint68u
As prerequisites, ensure you have Tapenade installed and gfortran/ifort compiler available and (for now until there is a fix) glibc version >= 2.34.
General instructions for installing Tapenade#
These insteructions are NOT for pcluster, but for other systems. The reason being that the glibc on pcluster is version 2.31 and not the 2.34 that is required by Tapenade.
We detail below the instructions for Linux, but the latest instructions for many operating systems can always be found here.
Before installing Tapenade, you must check that an up-to-date Java Runtime Environment is installed. Tapenade will not run with old Java Runtime Environments. Also, read the Tapenade license here.
Download the code from here into your chosen installation directory
install_dir.Go to your chosen installation directory
install_dir, and extract Tapenade from the tar file :
$ tar xvfz tapenade_3.16.tar
On Linux, depending on your distribution, Tapenade may require you to set the shell variable
JAVA_HOMEto your Java installation directory. It is oftenJAVA_HOME=/usr/java/default. You might also need to modify thePATHby adding the bin directory from the Tapenade installation. Every time you wish to use the AD capability with Tapenade, you must re-source the environment. We recommend that this be done automatically in your bash profile upon login. Exact commands will vary, but here’s an example of some lines in one.bashrc:
module load java/jdk/16.0.1 # Required by Tapenade, automatically sets JAVA_HOME in this particular case.
export TAPENADE_HOME="$HOME/tapenade_3.16"
export PATH="$PATH:$TAPENADE_HOME/bin"
You should now have a working copy of Tapenade. For more information on the
tapenadecommand and its arguments, type :
$ tapenade - ?
Instructions for pcluster#
Thanks to Ian, we finally have this working.
Open a singularity container that contains Tapenade as follows:
$ singularity shell --bind /efs_ecco:/efs_ecco /efs_ecco/singularity/tapenade/tapenade_v1.sif
Change the variable
JAVA_HOMEif Tapenade is not able to find Java (seems to be the case at least for me).
$ export JAVA_HOME=/usr
Tapenade should now work correctly inside the container. If you want to run an executable that was already compiled inside a container from pcluster i.e. outside the container, run the following command.
$ singularity exec --bind /efs_ecco:/efs_ecco /efs_ecco/singularity/tapenade/tapenade_v1.sif relative_path_to_executable
Running MITgcm-AD v2#
Running an adjoint simulation with MITgcm using Tapenade is not very different from running any other typical MITgcm simulation and broadly involves the same steps.
Go to the build directory of the
tutorial_global_oce_biogeoverification experiment. From the root of the MITgcm repository, this is the path:
$ cd verification/tutorial_global_oce_biogeo/build
Change the
nTimeStepsparameter ininput_ad/datafile based on how long you want the simulation to be.Clean any previous remnant files in your build subdirectory from a previous simulation, if any.
$ make CLEAN
Generate a makefile using the
genmake2script in the MITgcm. This command is run from the build subdirectory of your setup. The-tapoption specifies that this is a Tapenade-based adjoint simulation. Thecode_tapdirectory refers to the Tapenade-equivalent of thecode_addirectory used with TAF. Note that the optfile (option-of) can change based on the OS, compiler, and hardware. For the Singularity container that contains Tapenade, the optfile that works well istools/build_options/linux_amd64_gfortran.
$ ../../../tools/genmake2 -tap -of ../../../tools/build_options/linux_amd64_ifort -mods ../code_tap
Run the make commands.
tap_adjis the target name for the Tapenade adjoint mode andtap_tlmis the target name for the Tapenade tangent-linear mode. We will be using the adjoint mode here.
$ make depend
$ make -j 8 tap_adj
These commands will generate the adjoint/TLM executable mitgcmuv_tap_[adj,tlm].
In the future, when Tapenade is integrated with MITgcm diagnostics, one will be able to simply rely on these diagnostics to get the relevant fields such as temperature output in either binary or NetCDF format. Currently, one must instead do the I/O manually as shown below.
Open the Tapenade-generated file
the_main_loop_b.fwhich is a differentiated and preprocessed version of the original MITgcm filemodel/src/the_main_loop.Fand then add the following lines for I/O at the very end of the subroutine after all theDOloops but before the callsCALL POPREAL8ARRAY(theta,...)andCALL POPREAL8ARRAY(salt, ...)since these are calls related to the Tapenade tape and will erase the values in these fields (remember, the adjoint code goes in the reverse direction so the last values of these fields will be zeros).
open(unit=500, file='hFacC_biogeo.data')
open(unit=501, file='theta_biogeo.data')
open(unit=502, file='salt_biogeo.data')
DO ii1=1,nsy
DO ii2=1,nsx
DO ii3=1,nr
DO ii4=1-oly,oly+sny
DO ii5=1-olx,olx+snx
write(500,*) hFacC(ii5,ii4,ii3,ii2,ii1)
write(501,*) theta(ii5,ii4,ii3,ii2,ii1)
write(502,*) salt(ii5,ii4,ii3,ii2,ii1)
ENDDO
ENDDO
ENDDO
ENDDO
ENDDO
close(500)
close(501)
close(502)
Then add the following lines at the very end of the subroutine to get the adjoint values or the gradient with respect to the initial values of salt, theta.
open(unit=503, file='thetab_biogeo.data')
open(unit=504, file='saltb_biogeo.data')
DO ii1=1,nsy
DO ii2=1,nsx
DO ii3=1,nr
DO ii4=1-oly,oly+sny
DO ii5=1-olx,olx+snx
write(503,*) thetab(ii5,ii4,ii3,ii2,ii1)
write(504,*) saltb(ii5,ii4,ii3,ii2,ii1)
ENDDO
ENDDO
ENDDO
ENDDO
ENDDO
close(503)
close(504)
Copy this manually edited
the_main_loop_b.ffile for safekeeping and reuse since re-running the Tapenade command will generate a fresh file without the manual changes.
$ cp the_main_loop_b.f ..
In
genmake2, add the following line to the steps for targetTAP_ADJ_FILES. This will copy the safely kept, manually edited filethe_main_loop_b.fback to the build directory after the Tapenade command is run and before the final compilation so the manual changes have the desired effect.
$ cp ../the_main_loop_b.f .
Make the target
tap_[adj,tlm]again to incorporate I/O related changes in the executablemitgcmuv_tap_[adj,tlm].
$ make -j 8 tap_adj
Linking the input files and the executable in the run directory. This is a standard step in the MITgcm. It is performed from the run directory.
$ cd ../run
$ rm -r *
$ ln -s ../input_tap/* .
$ ../input_tap/prepare_run
$ ln -s ../build/mitgcmuv_tap_adj .
Running the executable and keeping logs.
$ ./mitgcmuv_tap_adj > output_tap_adj.txt 2>&1
One can analyze and plot the 2D fields after reading the output files, for example,
thetab_biogeo.dataand running it through the following Python commands:
thetab = np.loadtxt(thetab_biogeo.data)
# New shape from code_tap/SIZE.h
thetab = np.reshape(thetab, (2,2,15,40,72))
# Get rid of the halo points
thetab = thetab[:, :, z_level, 4:-4, 4:-4]
thetab_globe = np.zeros((64,128), dtype = float)
thetab_globe[:32,:64] = thetab[0,0,:,:]
thetab_globe[32:,:64] = thetab[1,0,:,:]
thetab_globe[:32,64:] = thetab[0,1,:,:]
thetab_globe[32:,64:] = thetab[1,1,:,:]
thetab_globe has the shape (64,128).
Get the water-land mask for the surface in a similar fashion by reading in
hFacC_biogeo.dataand running it through the same commands as above.
hfacc = np.loadtxt(hFacC_biogeo.data)
# New shape from code_tap/SIZE.h
hfacc = np.reshape(hfacc, (2,2,15,40,72))
# Get rid of the halo points
hfacc = hfacc[:, :, z_level, 4:-4, 4:-4]
hfacc_globe = np.zeros((64,128), dtype = float)
hfacc_globe[:32,:64] = hfacc[0,0,:,:]
hfacc_globe[32:,:64] = hfacc[1,0,:,:]
hfacc_globe[:32,64:] = hfacc[0,1,:,:]
hfacc_globe[32:,64:] = hfacc[1,1,:,:]
hfacc_globe also has the shape (64,128).
The filled contour plots in the paper from where we reference this tutorial are then created by plotting
hfacc_globe*thetab_globeto account for the land mask while plotting the adjoint of the temperature field.
Optional Matlab script#
The exact plots in the paper were actually generated using a Matlab script, given below for completeness. The script also requires TAF output for comparison with the Tapenade output.
% ------------------------------------------------------------------------------------------------------------------
% plot adxx for tutorial_global_oce_biogeo output used for Shreyas' 2024 JLESC paper
%
% LAST UPDATED: 9 Jan 2024
% ------------------------------------------------------------------------------------------------------------------
% --------------------
% set experiment names
% --------------------
inputs.rootdir='/scratch2/pillarh/MITgcm_nonECCO/';
inputs.exptname = 'tutorial_global_oce_biogeo/run_adj_Jan30_global_flux_CO2_1month_TAF_nchklev1is60_nogrdchk_nodiag/';
inputs.exptname_short = 'tutorial_global_oce_biogeo'; % needed for figure titles/names
inputs.fwdrunwithdiags = 'tutorial_global_oce_biogeo/run_tutorial_global_oce_biogeo_fwdwithdiags_nt60/';;
% -----------------------
% select controls to plot
% -----------------------
inputs.controls = {'taux','tauy','ustress','vstress','theta'}; % not sure what fu and fv are, but suspect momentum fluxes
%unitstr={'N/m2','N/m2','N/m2','N/m2','oC'};
%sssign=[1,1,1,1]; % need to switch sign of [tauu,tauv]?
inputs.controls_adxx = {'theta','fu','fv'}; % not sure what fu and fv are, but suspect momentum fluxes
%unitstr_adxx={'oC'};
% ------------------------------
% set up directories on Sverdrup
% ------------------------------
addpath '/home/pillarh/HP_MATLAB_SVERDRUP/HP_MATLAB_GENERIC'
addpath_recurse('/home/pillarh/HP_MATLAB_SVERDRUP/')
% ----------------------------------------------------------------------------------------------------------
% expt config
% ----------------------------------------------------------------------------------------------------------
inputs.rundir = [inputs.rootdir,inputs.exptname];
inputs.griddir = [inputs.rundir,'GRID/'];
inputs.figdir = [inputs.rundir,'FIGURES/'];
inputs.diagsdir = [inputs.rundir,'ADJfiles/'];
inputs.diagsdir_adxx = [inputs.rundir,'ADXXfiles/'];
if(exist(inputs.figdir)==0);mkdir(inputs.figdir);end;
inputs.nx = 128;
inputs.ny = 64;
inputs.nz = 15;
inputs.nIter0 = 5184000;
% ---------
% time info
% ---------
inputs.deltaT = 43200;
% ////////////////////////
% END OF HARD CODED STUFF
% ////////////////////////
% -----------
% pickup grid
% ------------
xc =rdmds([inputs.griddir 'XC']);
yc =rdmds([inputs.griddir 'YC']);
rc =rdmds([inputs.griddir 'RC']);
Depth =rdmds([inputs.griddir 'Depth']);
figure;
%m_coast('patch',[.7 .7 .7]);
m_proj('miller','lon',[0 360],'lat',[-78 78]); %('robinson','lon',[-330 30])
h = m_pcolor(xc,yc,Depth); set(h,'edgecolor','none'); colorbar
%m_coast('patch',[.7 .7 .7]);
m_grid('tickdir','out','ticklen',.001,'linestyle',':')
title('Bathymetry (m)'); print('-dpng',[inputs.figdir,'Depth.png'])
maskC = Depth; maskC(maskC~=0)=1;
% -----------------------
% plot adj sensitivities
% -----------------------
for jj = 1:length(inputs.controls)
disp(['plotting sensitivities (ADJ) for control = ',inputs.controls{jj}])
flist = dir([inputs.diagsdir,'ADJ',inputs.controls{jj},'*.data']);
for kk = 1:length(flist)
filename = [flist(kk).folder,'/',flist(kk).name];
idot = find(flist(kk).name=='.');
niter_tmp = flist(kk).name(idot(1)+1:idot(2)-1);
niter = sprintf('%03d',str2num(niter_tmp));
disp('==============================================')
disp('==============================================')
disp(['reading file = ',filename,'; niter = ',niter])
disp('==============================================')
disp('==============================================')
ADJ{jj}.data{kk} = rdmds(filename(1:end-5));
figure
m_proj('robinson','lon',[0 360],'lat',[-85 85]); %('robinson','lon',[-330 30])
if strcmp(inputs.controls{jj},'theta')~=1
if ~isempty(find(~isnan(replace(ADJ{jj}.data{kk},0,NaN))))
h = m_pcolor(xc,yc,replace(ADJ{jj}.data{kk},0,NaN)); set(h,'edgecolor','none'); colorbar
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
ADJ{jj}.abma{kk} = max(max(abs(ADJ{jj}.data{kk})));
set(gca,'clim',[-ADJ{jj}.abma{kk} ADJ{jj}.abma{kk}]);
h = title({['dJ/d',inputs.controls{jj},' at nIter = ',niter,' (t = ',num2str(str2num(niter)*inputs.deltaT/86400),' days)'],...
inputs.exptname_short});
else
disp(['No non-zero or non-nan values of sensitivity for control = ',inputs.controls{jj}])
end
else
data1 = ADJ{jj}.data{kk};
if ~isempty(find(~isnan(replace(data1(:,:,1),0,NaN))))
h = m_pcolor(xc,yc,replace(squeeze(data1(:,:,1)),0,NaN)); set(h,'edgecolor','none'); colorbar
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
ADJ{jj}.abma{kk} = max(max(abs(data1(:,:,1))));
set(gca,'clim',[-ADJ{jj}.abma{kk} ADJ{jj}.abma{kk}]);
h = title({['dJ/d',inputs.controls{jj},' at z = 25 m, nIter = ',...
niter,' (t = ',num2str(str2num(niter)*inputs.deltaT/86400),' days)'],inputs.exptname_short});
else
disp(['No non-zero or non-nan values of sensitivity for control = ',inputs.controls{jj}])
end
end
set(h,'interpreter','none');
hold on;
[c h] = m_contour(xc,yc,maskC,'-k'); set(h,'linewidth',1);
m_grid('box','fancy','tickdir','out','ticklen',.001,'xticklabels',[]);
figname = [inputs.figdir,'ADJ',inputs.controls{jj},'_t',niter,'.png']
print('-dpng',figname);
close all
end
end
return
% -------------------------------------------
% read Shreyas' initial condition sensitivity
% -------------------------------------------
filename = [inputs.rundir,'biogeo.nc'];
data_tap = ncread(filename,'thetab_biogeo_surf2D');
figure(100)
m_proj('robinson','lon',[0 360],'lat',[-85 85]); %('robinson','lon',[-330 30])
h = m_pcolor(xc,yc,replace(data_tap,0,NaN)); set(h,'edgecolor','none'); colorbar
tap.abma = max(max(abs(data_tap)));
set(gca,'clim',[-tap.abma tap.abma]);
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
%h1 = title({['Tapenade dJ/dtheta at t = 0, z = 25 m'],inputs.exptname_short})
h1 = title(['Tapenade dJ/dtheta at t = 0, z = 25 m'])
set(h1,'interpreter','none');
hold on;
[c h] = m_contour(xc,yc,maskC,'-k'); set(h,'linewidth',2);
m_grid('box','fancy','tickdir','out','ticklen',.001,'xticklabels',[]);
set(gca,'fontsize',12)
figname_tapenade = [inputs.figdir,'tapenade_adjtheta_t0.png'];
print('-dpng',figname_tapenade);
% will check I plot on same color range as TAF and then resave below
% ----------
% plot adxx
% ----------
for jj = 1:length(inputs.controls_adxx)
filename = [inputs.diagsdir_adxx,'adxx_',inputs.controls_adxx{jj},'.0000000000'];
disp('==============================================')
disp('==============================================')
disp(['reading file = ',filename])
disp('==============================================')
disp('==============================================')
adxx{jj}.data = rdmds(filename);
figure(100+jj)
m_proj('robinson','lon',[0 360],'lat',[-85 85]); %('robinson','lon',[-330 30])
if strcmp(inputs.controls_adxx{jj},'theta')~=1
if ~isempty(find(~isnan(replace(adxx{jj}.data,0,NaN))))
h = m_pcolor(xc,yc,replace(adxx{jj}.data,0,NaN)); set(h,'edgecolor','none'); colorbar
adxx{jj}.abma = max(max(abs(adxx{jj}.data)));
set(gca,'clim',[-adxx{jj}.abma adxx{jj}.abma]);
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
h1 = title({['dJ/d',inputs.controls_adxx{jj},' at t = 0'],inputs.exptname_short})
else
disp(['No non-zero or non-nan values of sensitivity for control = ',inputs.controls_adxx{jj}])
end
else
if ~isempty(find(~isnan(replace(adxx{jj}.data(:,:,1),0,NaN))))
h = m_pcolor(xc,yc,replace(squeeze(adxx{jj}.data(:,:,1)),0,NaN)); set(h,'edgecolor','none'); colorbar
adxx{jj}.abma = max(max(abs(adxx{jj}.data(:,:,1))));
set(gca,'clim',[-adxx{jj}.abma adxx{jj}.abma]);
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
%h1 = title({['dJ/d',inputs.controls_adxx{jj},' at t = 0, z = 25 m'],inputs.exptname_short})
h1 = title(['TAF dJ/d',inputs.controls_adxx{jj},' at t = 0, z = 25 m'])
else
disp(['No non-zero or non-nan values of sensitivity for control = ',inputs.controls_adxx{jj}])
end
end
set(h1,'interpreter','none');
hold on;
[c h] = m_contour(xc,yc,maskC,'-k'); set(h,'linewidth',2);
m_grid('box','fancy','tickdir','out','ticklen',.001,'xticklabels',[]);
figname_taf{jj} = [inputs.figdir,'adxx',inputs.controls_adxx{jj},'_t0.png'];
set(gca,'fontsize',12)
print('-dpng',figname_taf{jj});
end
% check taf and tapenade are saved on same colormap
itheta = find(strcmp(inputs.controls_adxx,'theta')==1);
if tap.abma>adxx{itheta}.abma
figure(100+itheta)
set(gca,'clim',[-tap.abma tap.abma]);
print('-dpng',figname_taf{itheta});
else
figure(100)
set(gca,'clim',[-adxx{jj}.abma adxx{jj}.abma]);
print('-dpng',figname_tapenade);
end
close all
% --------------------------------------
% plot their difference (Tapenade - TAF)
% --------------------------------------
figure
m_proj('robinson','lon',[0 360],'lat',[-85 85]); %('robinson','lon',[-330 30])
adj_diff = data_tap-squeeze(adxx{itheta}.data(:,:,1));
h = m_pcolor(xc,yc,replace(adj_diff,0,NaN)); set(h,'edgecolor','none'); colorbar
abma = max(max(abs(adj_diff)));
set(gca,'clim',[-abma abma]);
cmp=lbmap(30,'FrenchDiverging','reverse'); colormap(cmp);
h1 = title(['dJ/dtheta difference (Tapenade - TAF)'])
set(h1,'interpreter','none');
hold on;
[c h] = m_contour(xc,yc,maskC,'-k'); set(h,'linewidth',2);
m_grid('box','fancy','tickdir','out','ticklen',.001,'xticklabels',[]);
figname = [inputs.figdir,'Tapenade_minus_TAF_dJdtheta_t0.png'];
set(gca,'fontsize',12)
print('-dpng',figname);
% -------------------------------------
% plot air-sea CO2 flux and ocean pCO2
% from forward run, to try to understand
% sensitivity distribution
% -------------------------------------
diagname = {'DIC_pCO2tave','DIC_fluxCO2ave'}
%'DICCFLX';
varskip = [8,7]; % check meta to see which entries these are
units = {'atm','mol/m2/s'};
%flist = dir([inputs.rootdir,inputs.fwdrunwithdiags,'diags/surfDiag*.data']);
for jj = 1:length(diagname)
flist = dir([inputs.rootdir,inputs.fwdrunwithdiags,'diags/',diagname{jj},'*data']);
disp(['Plotting ',diagname{jj}])
for kk = 1:length(flist)
filename = [flist(kk).folder,'/',flist(kk).name];
idot = find(flist(kk).name=='.');
niter_tmp = flist(kk).name(idot(1)+1:idot(2)-1);
niter = sprintf('%03d',str2num(niter_tmp));
disp('==============================================')
disp('==============================================')
disp(['reading file = ',filename,'; niter = ',niter])
disp('==============================================')
disp('==============================================')
diags{jj}.data{kk} = rdmds(filename(1:end-5));
%diags{jj}.data{kk} = readbin(filename,[inputs.nx inputs.ny],1,'real*4',varskip(jj));
figure
m_proj('robinson','lon',[0 360],'lat',[-85 85]); %('robinson','lon',[-330 30])
numday = num2str((str2num(niter)-inputs.nIter0)*inputs.deltaT/86400)
if ~isempty(find(~isnan(replace(diags{jj}.data{kk},0,NaN))))
h = m_pcolor(xc,yc,replace(diags{jj}.data{kk},0,NaN)); set(h,'edgecolor','none'); colorbar
diags{jj}.abma{kk} = max(max(abs(diags{jj}.data{kk})));
diags{jj}.mi{kk} = min(min((diags{jj}.data{kk})));
diags{jj}.ma{kk} = max(max((diags{jj}.data{kk})));
if strcmp(diagname{jj},'DICCFLX')==1 || strcmp(diagname{jj},'DIC_fluxCO2ave')==1
cmp=lbmap(30,'FrenchDiverging','reverse');
set(gca,'clim',[-diags{jj}.abma{kk} diags{jj}.abma{kk}]);
else
cmp=lbmap(30,'samsmapwithwhitezero','reverse');
set(gca,'clim',[diags{jj}.mi{kk} diags{jj}.ma{kk}]);
end
colormap(cmp);
h1 = title({[diagname{jj},' (',units{jj},') at t = ',num2str(numday),' days'],...
inputs.exptname_short});
else
disp(['No non-zero or non-nan values of sensitivity for ',diagname{jj}])
end
set(h1,'interpreter','none');
hold on;
[c h] = m_contour(xc,yc,maskC,'-k'); set(h,'linewidth',2);
m_grid('box','fancy','tickdir','out','ticklen',.001,'xticklabels',[]);
figname = [inputs.figdir,diagname{jj},'_day_',numday,'.png'];
print('-dpng',figname);
close all
end
end
set(gca,'clim',[-tap.abma tap.abma]);
This script will allow you to generate this image:
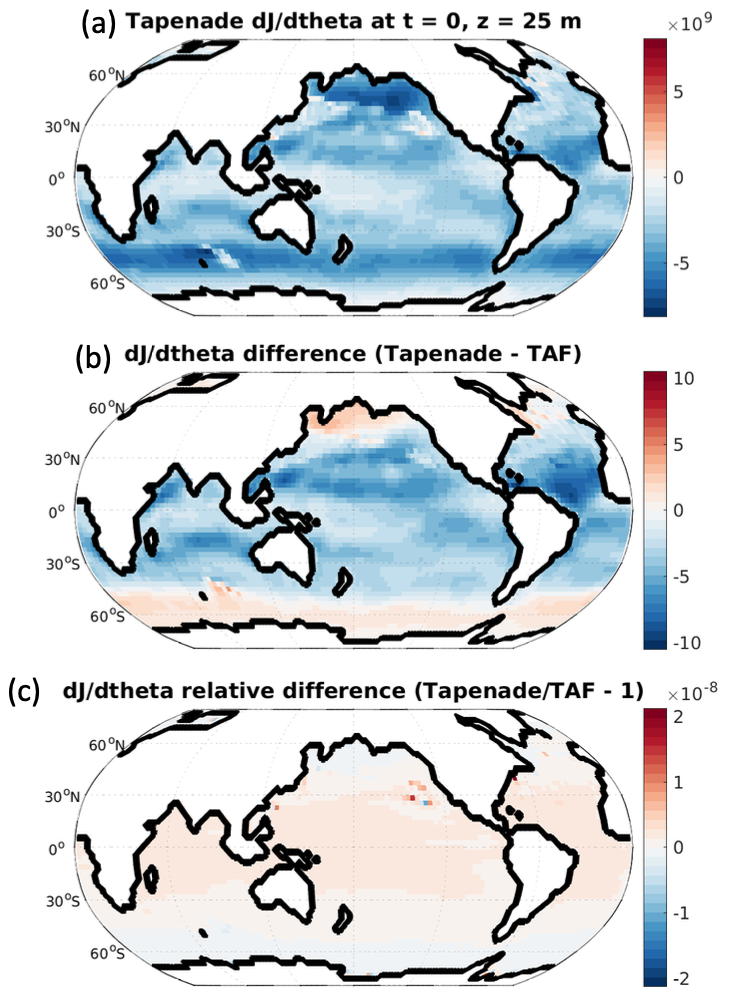
(a) Adjoint sensitivity (in mol/\(^\circ\)C) of the globally integrated air-sea flux of CO\(_2\) on the final day of the integration to the initial SST in the experiment tutorial_global_oce_biogeo, computed using Tapenade-generated code. (b) The difference (in mol/\(^\circ\)C) between sensitivity maps generated with Tapenade- versus TAF-generated adjoint code. Note the difference in color scale between (a) and (b), indicating the strong quantitative consistency between sensitivities calculated with the two AD tools. Their pointwise differences are 0(10\(^{8}\)) times smaller than the sensitivity amplitude itself (panel c).

