Example calculations with scalar quantities#
Objectives#
To demonstrate basic calculations using scalar fields (e.g., SSH, T, S) from the state estimate including: time series of mean quantities, spatial patterns of mean quantities, spatial patterns of linear trends, and spatial patterns of linear trends over different time periods.
Introduction#
We will demonstrate global calculations with SSH (global mean sea level time series, mean dynamic topography, global mean sea level trend) and a regional calculation with THETA (The Nino 3.4 index).
For this tutorial we will need the grid file, monthly SSH and THETA (potential temperature) spanning the years 1993 through 2017, and daily SSH for the year 1994. If you need to download these data from PO.DAAC, the ShortNames of the datasets are:
ECCO_L4_GEOMETRY_LLC0090GRID_V4R4
ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4 (1993-2017)
ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4 (1993-2017)
ECCO_L4_SSH_LLC0090GRID_DAILY_V4R4 (1994)
Global calculations with SSH#
First, load SSH and THETA fields and the model grid parameters.
import numpy as np
import sys
import xarray as xr
from copy import deepcopy
import matplotlib.pyplot as plt
%matplotlib inline
import glob
import warnings
warnings.filterwarnings('ignore')
# indicate mode of access
# options are:
# 'download': direct download from internet to your local machine
# 'download_ifspace': like download, but only proceeds
# if your machine have sufficient storage
# 's3_open': access datasets in-cloud from an AWS instance
# 's3_open_fsspec': use jsons generated with fsspec and
# kerchunk libraries to speed up in-cloud access
# 's3_get': direct download from S3 in-cloud to an AWS instance
# 's3_get_ifspace': like s3_get, but only proceeds if your instance
# has sufficient storage
access_mode = 's3_open_fsspec'
## Import the ecco_v4_py library into Python
## =========================================
## If ecco_v4_py is not installed in your local Python library,
## tell Python where to find it. The example below adds
## ecco_v4_py to the user's path if it is stored in the folder
## ECCOv4-py under the user's home directory
from os.path import join,expanduser
user_home_dir = expanduser('~')
sys.path.append(join(user_home_dir,'ECCOv4-py'))
import ecco_v4_py as ecco
import ecco_access as ea
## Set top-level file directory for the ECCO NetCDF files
## =================================================================
## currently set to ~/Downloads/ECCO_V4r4_PODAAC,
## the default if ecco_podaac_download was used to download dataset granules
ECCO_dir = join('/efs_ecco','ECCO_V4r4_PODAAC')
# for access_mode = 's3_open_fsspec', need to specify the root directory
# containing the jsons
jsons_root_dir = join('/efs_ecco','mzz-jsons')
## if working in the AWS cloud, access datasets needed for this tutorial
ShortNames_list = ["ECCO_L4_GEOMETRY_LLC0090GRID_V4R4",\
"ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4",\
"ECCO_L4_TEMP_SALINITY_LLC0090GRID_MONTHLY_V4R4"]
files_dict = ea.ecco_podaac_access(ShortNames_list,\
StartDate='1993-01',EndDate='2017-12',\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)
Now we’ll load daily SSH from 1994. We will do this in two ways. The first way creates a list of the files using glob, then loads each file into memory using open_dataset. The second way uses open_mfdataset which opens the files with the data variables as dask arrays first, then we load the dask arrays into memory. Note the difference in wall time (on a laptop):
## Load the model grid
if access_mode == 's3_open_fsspec':
ecco_grid = xr.open_dataset(files_dict[ShortNames_list[0]],\
engine='zarr',consolidated=False)
else:
ecco_grid = xr.open_dataset(files_dict[ShortNames_list[0]])
%%time
## 10 days of data without using dask (or the s3_open_fsspec access mode)
if access_mode == 's3_open_fsspec':
# don't use json (for comparison)
SSH_daily_files = ea.ecco_podaac_access(ShortNames_daily_list,\
StartDate='1994-01-01',EndDate='1994-01-10',\
mode='s3_open',\
jsons_root_dir=jsons_root_dir)\
[ShortNames_daily_list[0]]
else:
SSH_daily_files = files_SSH_daily_1994
def ds_load_concat(file_list,dim_concat,vars_to_drop):
for count,file in enumerate(file_list):
if count == 0:
ds_out = xr.open_dataset(file)
# remove variables not needed
ds_out = ds_out.drop_vars(vars_to_drop)
else:
ds_curr = xr.open_dataset(file)
# remove variables not needed
ds_curr = ds_curr.drop_vars(vars_to_drop)
# concatenate along dimension with name dim_concat
ds_out = xr.concat((ds_out,ds_curr),dim=dim_concat)
return ds_out
ds_SSH_daily_1994 = ds_load_concat(SSH_daily_files,dim_concat='time',\
vars_to_drop=['SSHNOIBC','SSHIBC','ETAN'])
ecco_daily_ds = xr.merge((ecco_grid,ds_SSH_daily_1994))
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
File <timed exec>:5
NameError: name 'ShortNames_daily_list' is not defined
%%time
## using dask
## (under the hood of the ea.ecco_podaac_to_xrdataset function)
ShortNames_daily_list = ["ECCO_L4_SSH_LLC0090GRID_DAILY_V4R4"]
ds_SSH_daily_1994 = ea.ecco_podaac_to_xrdataset(\
ShortNames_daily_list,\
StartDate='1994-01-01',EndDate='1994-12-31',\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)
ds_SSH_daily_1994 = ds_SSH_daily_1994.drop_vars(['SSHNOIBC','SSHIBC','ETAN'])
ecco_daily_ds = xr.merge((ecco_grid,ds_SSH_daily_1994)).compute()
CPU times: user 3.31 s, sys: 292 ms, total: 3.6 s
Wall time: 9.79 s
When in access_mode = s3_open_fsspec, the time difference in accessing data using json index/chunking files really becomes apparent. We could load a year of daily data using the json faster than we could load 10 days of data otherwise!
Now load monthly mean SSH, 1993-2017:
%%time
## Load monthly SSH and THETA data
ds_SSH_monthly = ea.ecco_podaac_to_xrdataset(\
ShortNames_list[1],\
StartDate='1993-01',EndDate='2017-12',\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)\
.drop_vars(['SSHNOIBC','SSHIBC','ETAN'])
CPU times: user 48 ms, sys: 4.01 ms, total: 52 ms
Wall time: 1.64 s
And monthly mean THETA, 1993-2017. But since these are larger files containing the full depth of the ocean, and we are subsetting the dataset (by removing the SALT variable that we don’t need), it is advantageous to use Dask here.
%%time
ds_THETA_monthly = ea.ecco_podaac_to_xrdataset(\
ShortNames_list[2],\
StartDate='1993-01',EndDate='2017-12',\
mode=access_mode,\
download_root_dir=ECCO_dir,\
max_avail_frac=0.5,\
jsons_root_dir=jsons_root_dir)\
.drop_vars('SALT')
## Merge the ecco_grid with the data variables to make ecco_monthly_ds
ecco_monthly_ds = xr.merge((ecco_grid,ds_SSH_monthly,ds_THETA_monthly))
CPU times: user 357 ms, sys: 60.7 ms, total: 418 ms
Wall time: 2.94 s
Display the first and last time entries in each dataset:
print(ecco_daily_ds.time[0].values)
print(ecco_daily_ds.time[-1].values)
print(ecco_monthly_ds.time[0].values)
print(ecco_monthly_ds.time[-1].values)
1994-01-01T12:00:00.000000000
1994-12-31T12:00:00.000000000
1993-01-16T12:00:00.000000000
2017-12-16T06:00:00.000000000
Sea surface height#
Global mean sea level#
Global mean sea surface height at time \(t\) is defined as follows:
Where \(SSH(i,t)\) is dynamic height at model grid cell \(i\) and time \(t\), \(A(i)\) is the area (m^2) of model grid cell \(i\)
There are several ways of doing the above calculations. Since this is the first tutorial with actual calcuations, we’ll present a few different approaches for getting to the same answer.
Part 1: \(A_{\text{global ocean}}\)#
Let’s start on the simplest quantity, the global ocean surface area \(A_{\text{global ocean}}\). Our calculation uses SSH which is a ‘c’ point variable. The surface area of tracer grid cells is provided by the model grid parameter rA. rA is a two-dimensional field that is defined over all model grid points, including land.
To calculate the total ocean surface area we need to ignore the area contributions from land.
We will first construct a 3D mask that is True for model grid cells that are wet and False for model grid cells that are dry cells.
# ocean_mask is ceiling of hFacC which is 0 for 100% dry cells and
# 0 > hFacC >= 1 for grid cells that are at least partially wet
# hFacC is the fraction of the thickness (h) of the grid cell which
# is wet. we'll consider all hFacC > 0 as being a wet grid cell
# and so we use the 'ceiling' function to make all values > 0 equal to 1.
ocean_mask = np.ceil(ecco_monthly_ds.hFacC)
ocean_mask = ocean_mask.where(ocean_mask==1, np.nan)
# print the dimensions and size of ocean_mask
print(type(ocean_mask))
print((ocean_mask.dims))
<class 'xarray.core.dataarray.DataArray'>
('k', 'tile', 'j', 'i')
plt.figure(figsize=(12,6), dpi= 90)
ecco.plot_tiles(ocean_mask.isel(k=0),layout='latlon', rotate_to_latlon=True)
# select out the model depth at k=1, round the number and convert to string.
z = str((np.round(ecco_monthly_ds.Z.values[0])))
plt.suptitle('Wet (1) /dry (0) mask for k=' + str(0) + ', z=' + z + 'm');
<Figure size 1080x540 with 0 Axes>
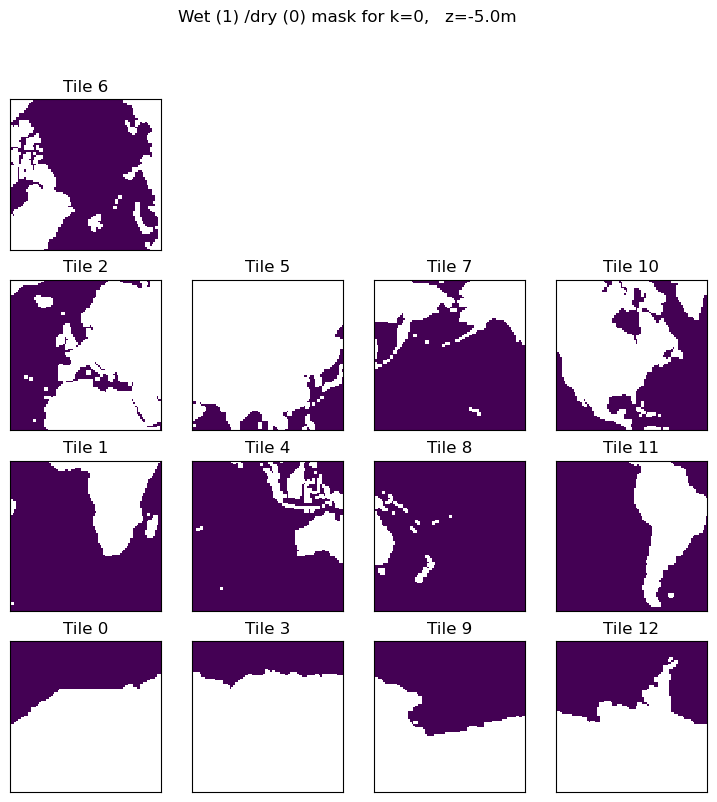
To calculate \(A_{\text{global ocean}}\) we must apply the surface wet/dry mask to \(rA\).
# Method 1: the array index method, []
# select land_c at k index 0
total_ocean_area = np.sum(ecco_monthly_ds.rA*ocean_mask[0,:])
# these three methods give the same numerical result. Here are
# three alternative ways of printing the result
print ('total ocean surface area ( m^2) %d ' % total_ocean_area.values)
print ('total ocean surface area (km^2) %d ' % (total_ocean_area.values/1.0e6))
# or in scientific notation with 2 decimal points
print ('total ocean surface area (km^2) %.2E' % (total_ocean_area.values/1.0e6))
total ocean surface area ( m^2) 358013844062208
total ocean surface area (km^2) 358013844
total ocean surface area (km^2) 3.58E+08
This compares favorably with approx 3.60 x 10\(^8\) km\(^2\) from https://hypertextbook.com/facts/1997/EricCheng.shtml
Multiplication of DataArrays#
You probably noticed that the multiplication of grid cell area with the land mask was done element by element. One useful feature of DataArrays is that their dimensions are automatically lined up when doing binary operations. Also, because rA and ocean_mask are both DataArrays, their inner product and their sums are also DataArrays.
Note:: ocean_mask has a depth (k) dimension while rA does not (horizontal model grid cell area does not change as a function of depth in ECCOv4). As a result, when rA is multiplied with ocean_mask
xarraybroadcasts rA to all k levels. The resulting matrix inherits the k dimension from ocean_mask.
Another way of summing over numpy arrays#
As rA and ocean both store numpy arrays, you can also calculate the sum of their product by invoking the .sum() command inherited in all numpy arrays:
total_ocean_area = (ecco_monthly_ds.rA*ocean_mask).isel(k=0).sum()
print ('total ocean surface area (km^2) ' + '%.2E' % (total_ocean_area.values/1e6))
total ocean surface area (km^2) 3.58E+08
Part2 : \(SSH_{\text{global mean}}(t)\)#
The global mean SSH at each \(t\) is given by,
One way of calculating this is to take advantage of DataArray coordinate labels and use its .sum() functionality to explicitly specify which dimensions to sum over:
# note no need to multiple RAC by land_c because SSH is nan over land
SSH_global_mean_mon = (ecco_monthly_ds.SSH*ecco_monthly_ds.rA).sum(dim=['i','j','tile'])/total_ocean_area
# remove time mean from time series
SSH_global_mean_mon = SSH_global_mean_mon-SSH_global_mean_mon.mean(dim='time')
# add helpful unit label
SSH_global_mean_mon.attrs['units']='m'
# and plot for fun
SSH_global_mean_mon.plot(color='k');plt.grid()
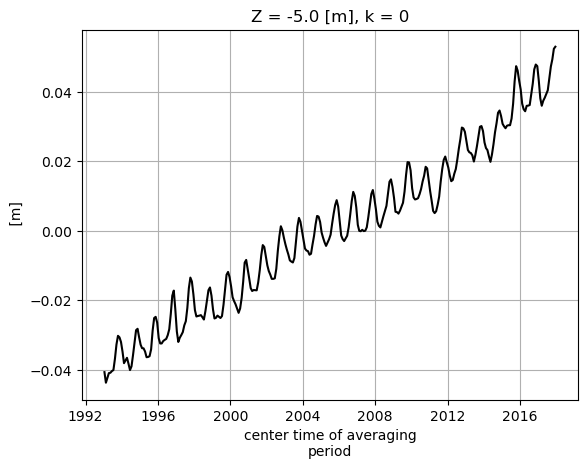
Alternatively we can do the summation over the three non-time dimensions. The time dimension of SSH is along the first dimension (axis) of the array, axis 0.
# note no need to multiple RAC by land_c because SSH is nan over land
SSH_global_mean = np.sum(ecco_monthly_ds.SSH*ecco_monthly_ds.rA,axis=(1,2,3))/total_ocean_area
SSH_global_mean = SSH_global_mean.compute()
Even though SSH has 3 dimensions (time, tile, j, i) and rA and ocean_mask.isel(k=0) have 2 (j,i), we can mulitply them. With xarray the element-by-element multiplication occurs over their common dimension.
The resulting \(SSH_{global-mean}\) DataArray has a single dimension, time.
Part 3 : Plotting the global mean sea level time series:#
Before we plot the global mean sea level curve let’s remove its time-mean to make it global mean sea level anomaly (the absolute value has no meaning here anyway).
plt.figure(figsize=(8,4), dpi= 90)
# Method 1: .mean() method of `DataArrays`
SSH_global_mean_anomaly = SSH_global_mean - SSH_global_mean.mean()
# Method 2: numpy's `mean` method
SSH_global_mean_anomaly = SSH_global_mean - np.mean(SSH_global_mean)
SSH_global_mean_anomaly.plot()
plt.grid()
plt.title('ECCO v4r4 Global Mean Sea Level Anomaly');
plt.ylabel('m');
plt.xlabel('year');
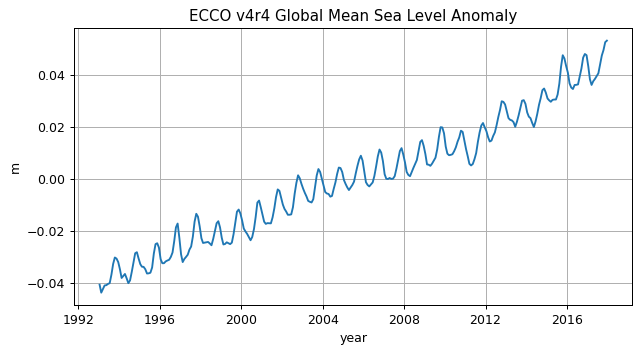
Mean Dynamic Topography#
Mean dynamic topography is calculated as follows,
Where \(nt\) is the number of time records.
For MDT we preserve the spatial dimensions. Summation and averaging are over the time dimensions (axis 0).
## Two equivalent methods
# Method 1, specify the axis over which to average
MDT = np.mean(ecco_monthly_ds.SSH - SSH_global_mean,axis=0)
# Method 2, specify the coordinate label over which to average
MDT_B = (ecco_monthly_ds.SSH - SSH_global_mean).mean(dim=['time'])
# which can be verified using the '.equals()' method to compare Datasets and DataArrays
print(MDT.equals(MDT_B))
True
As expected, MDT has preserved its spatial dimensions:
MDT.dims
('tile', 'j', 'i')
Before plotting the MDT field remove its spatial mean since its spatial mean conveys no dynamically useful information.
MDT_no_spatial_mean = MDT - MDT*ecco_monthly_ds.rA/total_ocean_area
MDT_no_spatial_mean.shape
(13, 90, 90)
plt.figure(figsize=(12,6), dpi= 90)
# mask land points to Nan
MDT_no_spatial_mean = MDT_no_spatial_mean.where(ocean_mask[0,:] !=0)
ecco.plot_proj_to_latlon_grid(ecco_monthly_ds.XC, \
ecco_monthly_ds.YC, \
MDT_no_spatial_mean*ocean_mask.isel(k=0), \
user_lon_0=0,\
plot_type = 'pcolormesh', \
cmap='seismic',show_colorbar=True,\
dx=2,dy=2);
plt.title('ECCO v4r4 Mean Dynamic Topography [m]');
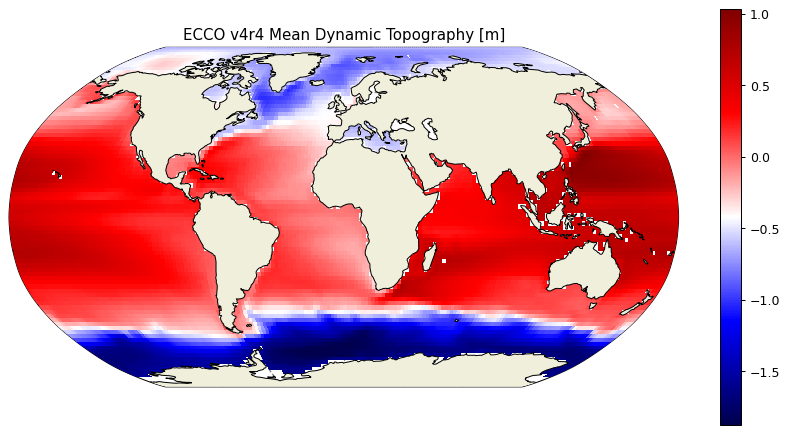
Spatial variations of sea level linear trends#
To calculate the linear trend for the each model point we will use on the polyfit function of numpy. First, define a time variable in years for SSH.
days_since_first_record = ((ecco_monthly_ds.time - ecco_monthly_ds.time[0])/(86400e9)).astype(int).values
len(days_since_first_record)
300
Next, reshape the four dimensional SSH field into two dimensions, time and space (t, i)
ssh_flat = np.reshape(ecco_monthly_ds.SSH.values, (len(ecco_monthly_ds.SSH.time), 13*90*90))
ssh_flat.shape
(300, 105300)
Now set all SSH values that are ‘nan’ to zero because the polynominal fitting routine can’t handle nans,
ssh_flat[np.nonzero(np.isnan(ssh_flat))] = 0
ssh_flat.shape
(300, 105300)
Do the polynomial fitting, https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.polyfit.html
# slope is in m / day
ssh_slope, ssh_intercept = np.polyfit(days_since_first_record, ssh_flat, 1)
print(ssh_slope.shape)
# and reshape the slope result back to 13x90x90
ssh_slope = np.reshape(ssh_slope, (13, 90,90))
# mask
ssh_slope_masked = np.where(ocean_mask[0,:] > 0, ssh_slope, np.nan)
# convert from m / day to mm/year
ssh_slope_mm_year = ssh_slope_masked*365*1e3
(105300,)
plt.figure(figsize=(12,6), dpi= 90)
ecco.plot_proj_to_latlon_grid(ecco_monthly_ds.XC, \
ecco_monthly_ds.YC, \
ssh_slope_mm_year, \
user_lon_0=-66,\
plot_type = 'pcolormesh', \
show_colorbar=True,\
dx=1, dy=1, cmin=-8, cmax=8)
plt.title('ECCO v4r4 Sea Level Trend [mm/yr]');
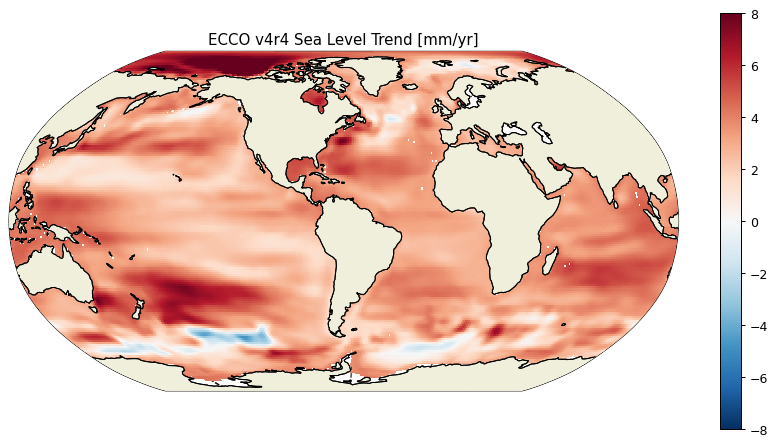
And the mean rate of global sea level change in mm/year over the 1993-2017 period is:
((ssh_slope_mm_year*ecco_monthly_ds.rA).sum())/((ecco_monthly_ds.rA*ocean_mask.isel(k=0)).sum())
<xarray.DataArray ()> Size: 8B
array(3.21794046)
Coordinates:
Z float32 4B -5.0
k int32 4B 0Constructing Monthly means from Daily means#
We can also construct our own monthly means from the daily means using this command: (See http://xarray.pydata.org/en/stable/generated/xarray.Dataset.resample.html for more information)
# note no need to multiple RAC by land_c because SSH is nan over land
SSH_global_mean_day = (ecco_daily_ds.SSH*ecco_daily_ds.rA).sum(dim=['i','j','tile'])/total_ocean_area
# remove time mean from time series and replace with 1994 time mean from monthly SSH
SSH_global_mean_day = SSH_global_mean_day-SSH_global_mean_day.mean(dim='time')\
+SSH_global_mean_mon.sel(time='1994').mean(dim='time')
# add helpful unit label
SSH_global_mean_day.attrs['units']='m'
# and plot for fun
SSH_global_mean_day.plot(color='k');plt.grid()
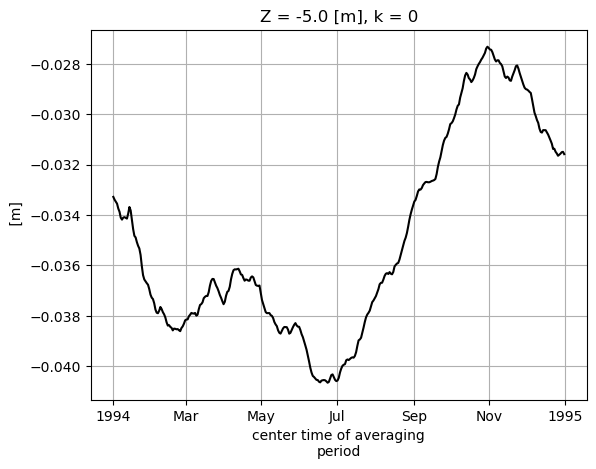
SSH_global_mean_mon_alt = SSH_global_mean_day.resample(time='1M', loffset='-15D').mean()
Plot to compare.
SSH_global_mean_mon.sel(time='1994').plot(color='r', marker='.');
SSH_global_mean_mon_alt.sel(time='1994').plot(color='g', marker='o');
plt.grid()
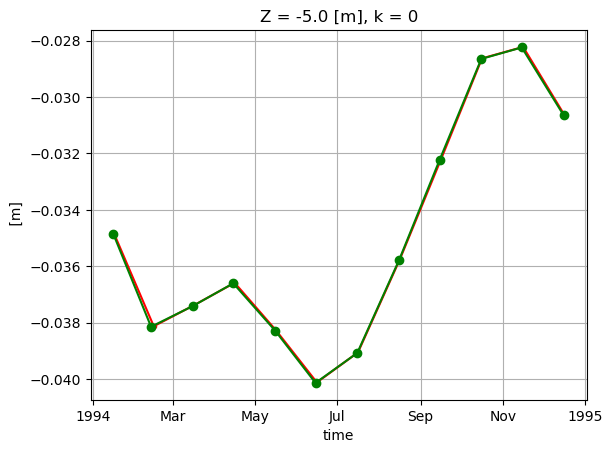
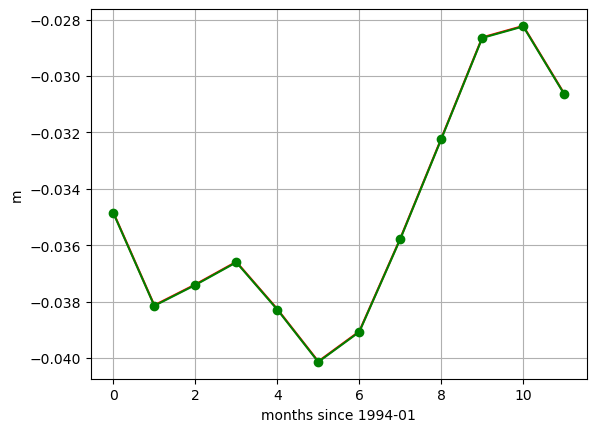
These small differences are simply an artifact of the time indexing. We used loffset=’15D’ to shift the time of the monthly mean SSH back 15 days, close to the center of the month. The SSH_global_mean_mon field is centered exactly at the middle of the month, and since months aren’t exactly 30 days long, this results in a small discrepancy when plotting with a time x-axis. If we plot without a time object x axis we find the values to be the same. That’s because ECCO monthly means are calculated over calendar months.
print ('date in middle of month')
print(SSH_global_mean_mon.time.values[0:2])
print ('\ndate with 15 day offset from the end of the month')
print(SSH_global_mean_mon_alt.time.values[0:2])
date in middle of month
['1993-01-16T12:00:00.000000000' '1993-02-15T00:00:00.000000000']
date with 15 day offset from the end of the month
['1994-01-16T00:00:00.000000000' '1994-02-13T00:00:00.000000000']
plt.plot(SSH_global_mean_mon.sel(time='1994').values, color='r', marker='.');
plt.plot(SSH_global_mean_mon_alt.sel(time='1994').values, color='g', marker='o');
plt.xlabel('months since 1994-01');
plt.ylabel('m')
plt.grid()
Regional calculations with THETA#
lat_bounds = np.logical_and(ecco_monthly_ds.YC >= -5, ecco_monthly_ds.YC <= 5)
lon_bounds = np.logical_and(ecco_monthly_ds.XC >= -170, ecco_monthly_ds.XC <= -120)
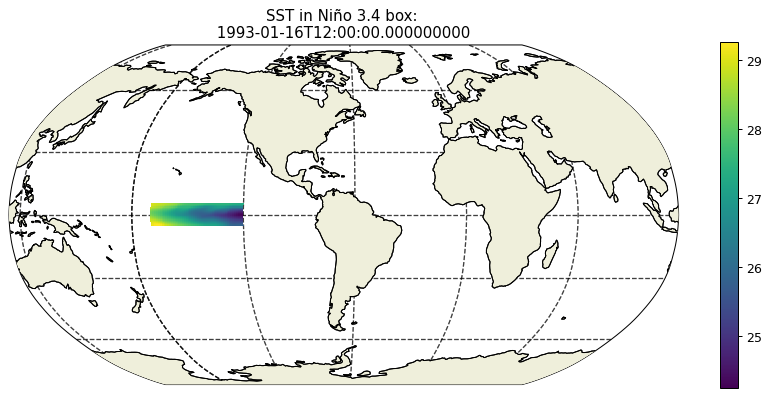
SST = ecco_monthly_ds.THETA.isel(k=0)
SST_masked=SST.where(np.logical_and(lat_bounds, lon_bounds))
plt.figure(figsize=(12,5), dpi= 90)
ecco.plot_proj_to_latlon_grid(ecco_monthly_ds.XC, \
ecco_monthly_ds.YC, \
SST_masked.isel(time=0),\
user_lon_0 = -66,\
show_colorbar=True);
plt.title('SST in Niño 3.4 box: \n %s ' % str(ecco_monthly_ds.time[0].values));
# Create the same mask for the grid cell area
rA_masked=ecco_monthly_ds.rA.where(np.logical_and(lat_bounds, lon_bounds));
# Calculate the area-weighted mean in the box
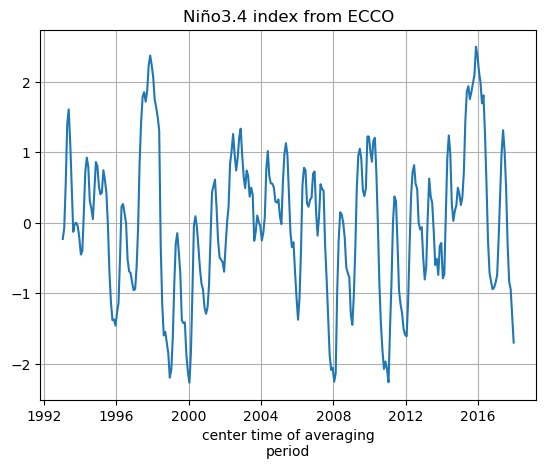
SST_masked_mean=(SST_masked*rA_masked).sum(dim=['tile','j','i'])/np.sum(rA_masked)
# Substract the temporal mean from the area-weighted mean to get a time series, the Nino 3.4 index
SST_nino_34_anom_ECCO_monthly_mean = SST_masked_mean - np.mean(SST_masked_mean)
Load up the Niño 3.4 index values from ESRL#
# https://psl.noaa.gov/gcos_wgsp/Timeseries/Data/nino34.long.anom.data
# NINO34
# 5N-5S 170W-120W
# HadISST
# Anomaly from 1981-2010
# units=degC
import urllib.request
data = urllib.request.urlopen('https://psl.noaa.gov/gcos_wgsp/Timeseries/Data/nino34.long.anom.data')
# the following code parses the ESRL text file and puts monthly-mean nino 3.4 values into an array
start_year = 1993
end_year = 2017
num_years = end_year-start_year+1
nino34_noaa = np.zeros((num_years, 12))
for i,l in enumerate(data):
line_str = str(l, "utf-8")
x=line_str.split()
try:
year = int(x[0])
row_i = year-start_year
if row_i >= 0 and year <= end_year:
print('loading Niño 3.4 for year %s row %s' % (year, row_i))
for m in range(0,12):
nino34_noaa[row_i, m] = float(x[m+1])
except:
continue
loading Niño 3.4 for year 1993 row 0
loading Niño 3.4 for year 1994 row 1
loading Niño 3.4 for year 1995 row 2
loading Niño 3.4 for year 1996 row 3
loading Niño 3.4 for year 1997 row 4
loading Niño 3.4 for year 1998 row 5
loading Niño 3.4 for year 1999 row 6
loading Niño 3.4 for year 2000 row 7
loading Niño 3.4 for year 2001 row 8
loading Niño 3.4 for year 2002 row 9
loading Niño 3.4 for year 2003 row 10
loading Niño 3.4 for year 2004 row 11
loading Niño 3.4 for year 2005 row 12
loading Niño 3.4 for year 2006 row 13
loading Niño 3.4 for year 2007 row 14
loading Niño 3.4 for year 2008 row 15
loading Niño 3.4 for year 2009 row 16
loading Niño 3.4 for year 2010 row 17
loading Niño 3.4 for year 2011 row 18
loading Niño 3.4 for year 2012 row 19
loading Niño 3.4 for year 2013 row 20
loading Niño 3.4 for year 2014 row 21
loading Niño 3.4 for year 2015 row 22
loading Niño 3.4 for year 2016 row 23
loading Niño 3.4 for year 2017 row 24
SST_nino_34_anom_ECCO_monthly_mean.plot();plt.grid()
plt.title('Niño3.4 index from ECCO')
Text(0.5, 1.0, 'Niño3.4 index from ECCO')
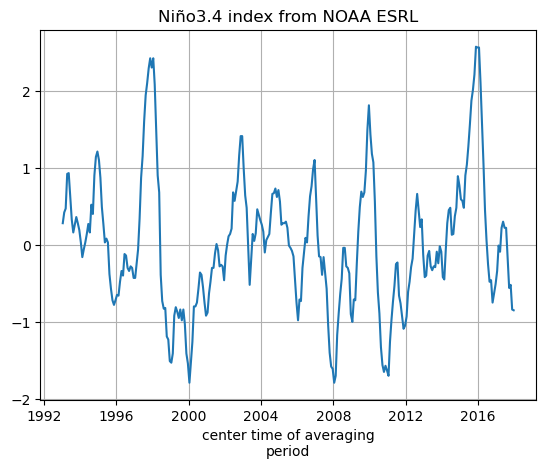
we’ll make a new DataArray for the NOAA SST nino_34 data by copying the DataArray for the ECCO SST data and replacing the values
SST_nino_34_anom_NOAA_monthly_mean = xr.DataArray(data=nino34_noaa.ravel(),\
dims=SST_nino_34_anom_ECCO_monthly_mean.dims,\
coords=SST_nino_34_anom_ECCO_monthly_mean.coords)
SST_nino_34_anom_NOAA_monthly_mean.plot();plt.grid()
plt.title('Niño3.4 index from NOAA ESRL')
Text(0.5, 1.0, 'Niño3.4 index from NOAA ESRL')
Plot the ECCOv4r4 and ESRL Niño 3.4 index#
# calculate correlation between time series
nino_corr = np.corrcoef(SST_nino_34_anom_ECCO_monthly_mean, SST_nino_34_anom_NOAA_monthly_mean)[1]
nino_ev = 1 - np.var(SST_nino_34_anom_ECCO_monthly_mean-SST_nino_34_anom_NOAA_monthly_mean)/np.var(SST_nino_34_anom_NOAA_monthly_mean)
plt.figure(figsize=(8,5), dpi= 90)
plt.plot(SST_nino_34_anom_ECCO_monthly_mean.time, \
SST_nino_34_anom_ECCO_monthly_mean - SST_nino_34_anom_ECCO_monthly_mean.mean(),'b.-')
plt.plot(SST_nino_34_anom_NOAA_monthly_mean.time, \
SST_nino_34_anom_NOAA_monthly_mean - SST_nino_34_anom_NOAA_monthly_mean.mean(),'r.-')
plt.title('Niño 3.4 SST Anomaly \n correlation: %s \n explained variance: %s' % (np.round(nino_corr[0],3), \
np.round(nino_ev.values,3)));
plt.legend(('ECCO','NOAA'))
plt.ylabel('deg C');
plt.xlabel('year');
plt.grid()
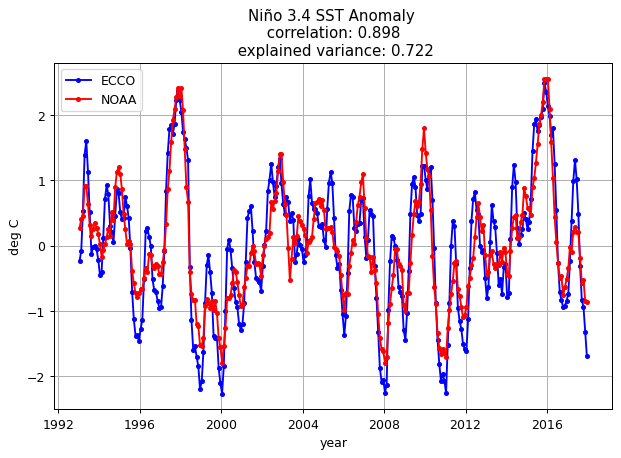
ECCO is able to match the NOAA Niño 3.4 index fairly well.
Conclusion#
You should now be familiar with doing some doing calculations using scalar quantities.
Suggested exercises#
Create the global mean SSH time series from 1992-2015
Create the global mean sea level trend (map) from 1992-2015
Create the global mean sea level trend (map) for two epochs 1992-2003, 2003-2015
Compare other Niño indices

