The ecco_access “package”: a starting point for accessing ECCO output on PO.DAAC#
Andrew Delman, updated 2024-10-13
Introduction
Add ecco_access to your Python path
Using the ecco_podaac_to_xrdataset function
Using the ecco_podaac_access function
Introduction#
In the past several years since ECCOv4 release 4 output was made available on the Physical Oceanography Distributed Active Archive Center or PO.DAAC, a number of Python scripts/functions have been written to facilitate requests of this output, authored by Jack McNelis, Ian Fenty, and Andrew Delman. To make access easier and standardize the format of these requests, the ecco_access library has been made available in the ecco_access folder of the ECCO-v4-Python-Tutorial Github repository.
This library I am calling a “package” in quotes because it currently has the core structure of any package you would install using conda or pip; there is an __init__.py file that allows you to access all of the library’s modules and the functions within, using a single import ecco_access command. However, this “package” is not available through conda or pip yet. For the convenience of the ECCO Hackathon, ecco_access has been copied over to the ecco-2024 repo used in the hackathon, with symbolic links included in the existing tutorial directories so that these tutorials can immediately use the library.
This tutorial will help you get set up with ecco_access so that you can use it from your tutorial directories on OSS or wherever else you need it. It also introduces the two top-level functions that you would likely use in ecco_access:
ecco_podaac_to_xrdataset: takes as input a text query or ECCO dataset identifier, and returns an xarray Datasetecco_podaac_access: takes the same input, but returns the URLs/paths or local files where the data is located
Add ecco_access to your Python path#
For more extensive use of this “package” (and sharing any edits with the community), I recommend cloning the ECCO-v4-Python-Tutorial repo and then adding the ecco_access folder to your Python path, using the steps below.
Clone the ECCO-v4-Python-Tutorial repository#
Navigate to your home directory before cloning the ECCO-v4-Python-Tutorial repo. This way the repo will appear as a directory under your home directory and is easily accessed.
cd ~
(notebook) jovyan@jupyter-adelman:~$ git clone git@github.com:ECCO-GROUP/ECCO-v4-Python-Tutorial.git
Add symlink pointing from your path#
We need to put the ecco_access folder from this repository into the search path for Python packages. There are at least two ways to do this. If you will have most or all of your Python-based notebooks and codes in one directory, you can create a symbolic link (symlink) from that directory (which is in the search path by default) to ecco_access. For instance, if the directory containing your codes is ~/working_repo/code_dir:
(notebook) jovyan@jupyter-adelman:~$ ln -s ~/ECCO-v4-Python-Tutorial/ecco_access ~/working_repo/code_dir
Alternatively, you can have the symlink point from a directory that is in the standard Python path; to identify these directories use python -c "import sys; print(sys.path)":
(notebook) jovyan@jupyter-adelman:~$ python -c "import sys; print(sys.path)"
['', '/srv/conda/envs/notebook/lib/python311.zip', '/srv/conda/envs/notebook/lib/python3.11',
'/srv/conda/envs/notebook/lib/python3.11/lib-dynload', '/srv/conda/envs/notebook/lib/python3.11/site-packages']
We’ll use the last directory listed in the path:
(notebook) jovyan@jupyter-adelman:~$ ln -s ~/ECCO-v4-Python-Tutorial/ecco_access /srv/conda/envs/notebook/lib/python3.11/site-packages/ecco_access
Using the ecco_podaac_to_xrdataset function#
Perhaps the most convenient way to use ecco_access is the ecco_podaac_to_xrdataset; it takes as input a query consisting of NASA Earthdata dataset ShortName(s), ECCO variables, or text strings in the variable descriptions, and outputs an xarray Dataset. Let’s look at the syntax:
import numpy as np
import xarray as xr
from os.path import join,expanduser
import matplotlib.pyplot as plt
import ecco_access as ea
# identify user's home directory
user_home_dir = expanduser('~')
help(ea.ecco_podaac_to_xrdataset)
Help on function ecco_podaac_to_xrdataset in module ecco_access.ecco_access:
ecco_podaac_to_xrdataset(query, version='v4r4', grid=None, time_res='all', StartDate=None, EndDate=None,\
snapshot_interval=None, mode='download_ifspace', download_root_dir=None, **kwargs)
This function queries and accesses ECCO datasets from PO.DAAC. The core query and download functions are adapted from Jupyter notebooks
created by Jack McNelis and Ian Fenty
(https://github.com/ECCO-GROUP/ECCO-ACCESS/blob/master/PODAAC/Downloading_ECCO_datasets_from_PODAAC/README.md)
and modified by Andrew Delman (https://ecco-v4-python-tutorial.readthedocs.io).
It is similar to ecco_podaac_access, except instead of a list of URLs or files,
an xarray Dataset with all of the queried ECCO datasets is returned.
Parameters
----------
query: str, list, or dict, defines datasets or variables to access.
If query is str, it specifies either a dataset ShortName (which is
assumed if the string begins with 'ECCO_'), or a text string that
can be used to search the ShortNames, variable names, and descriptions.
A query may also be a list of multiple ShortNames and/or text searches,
or a dict that contains grid,time_res specifiers as keys and ShortNames
or text searches as values, e.g.,
{'native,monthly':['ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4',
'THETA']}
will query the native grid monthly SSH datasets, and all native grid
monthly datasets with variables or descriptions matching 'THETA'.
version: ('v4r4'), specifies ECCO version to query
grid: ('native','latlon',None), specifies whether to query datasets with output
on the native grid or the interpolated lat/lon grid.
The default None will query both types of grids, unless specified
otherwise in a query dict (e.g., the example above).
time_res: ('monthly','daily','snapshot','all'), specifies which time resolution
to include in query and downloads. 'all' includes all time resolutions,
and datasets that have no time dimension, such as the grid parameter
and mixing coefficient datasets.
StartDate,EndDate: str, in 'YYYY', 'YYYY-MM', or 'YYYY-MM-DD' format,
define date range [StartDate,EndDate] for download.
EndDate is included in the time range (unlike typical Python ranges).
Full ECCOv4r4 date range (default) is '1992-01-01' to '2017-12-31'.
For 'SNAPSHOT' datasets, an additional day is added to EndDate to enable closed budgets
within the specified date range.
snapshot_interval: ('monthly', 'daily', or None), if snapshot datasets are included in ShortNames,
this determines whether snapshots are included for only the beginning/end of each month
('monthly'), or for every day ('daily').
If None or not specified, defaults to 'daily' if any daily mean ShortNames are included
and 'monthly' otherwise.
mode: str, one of the following:
'download': Download datasets using NASA Earthdata URLs
'download_ifspace': Check storage availability before downloading.
Download only if storage footprint of downloads
<= max_avail_frac*(available storage)
'download_subset': Download spatial and temporal subsets of datasets
via Opendap; query help(ecco_access.ecco_podaac_download_subset)
to see keyword arguments that can be used in this mode.
The following modes work within the AWS cloud only:
's3_open': Access datasets on S3 without downloading.
's3_open_fsspec': Use json files (generated with `fsspec` and `kerchunk`)
for expedited opening of datasets.
's3_get': Download from S3 (to AWS EC2 instance).
's3_get_ifspace': Check storage availability before downloading;
download if storage footprint
<= max_avail_frac*(available storage).
Otherwise data are opened "remotely" from S3 bucket.
download_root_dir: str, defines parent directory to download files to.
Files will be downloaded to directory download_root_dir/ShortName/.
If not specified, parent directory defaults to '~/Downloads/ECCO_V4r4_PODAAC/'.
Additional keyword arguments*:
*This is not an exhaustive list, especially for
'download_subset' mode; use help(ecco_access.ecco_podaac_download_subset) to display
options specific to that mode
max_avail_frac: float, maximum fraction of remaining available disk space to
use in storing ECCO datasets.
If storing the datasets exceeds this fraction, an error is returned.
Valid range is [0,0.9]. If number provided is outside this range, it is replaced by the closer
endpoint of the range.
jsons_root_dir: str, for s3_open_fsspec mode only, the root/parent directory where the
fsspec/kerchunk-generated jsons are found.
jsons are generated using the steps described here:
https://medium.com/pangeo/fake-it-until-you-make-it-reading-goes-netcdf4-data-on-aws-s3-as-zarr
-for-rapid-data-access-61e33f8fe685
and stored as {jsons_root_dir}/MZZ_{GRIDTYPE}_{TIME_RES}/{SHORTNAME}.json.
For v4r4, GRIDTYPE is '05DEG' or 'LLC0090GRID'.
TIME_RES is one of: ('MONTHLY','DAILY','SNAPSHOT','GEOMETRY','MIXING_COEFFS').
n_workers: int, number of workers to use in concurrent downloads. Benefits typically taper off above 5-6.
force_redownload: bool, if True, existing files will be redownloaded and replaced;
if False (default), existing files will not be replaced.
Returns
-------
ds_out: xarray Dataset or dict of xarray Datasets (with ShortNames as keys),
containing all of the accessed datasets.
This function does not work with the query modes: 'ls','query','s3_ls','s3_query'.
There are a lot of options that you can use to “submit” a query with this function. Let’s consider a simple case, where we already have the ShortName for the monthly native grid SSH from ECCOv4r4 (ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4), and we want to access output from the year 2017. The ShortName goes in the query field, and we can specify start and end dates (in YYYY-MM or YYYY-MM-DD format). The other options that matter most for this request are the mode, and depending on the mode, the download_root_dir or the jsons_root_dir.
Direct download over the internet (mode = ‘download’)#
Let’s try the download mode, which retrieves the data over the Internet using NASA Earthdata URLs (this should work on any machine with Internet access, including cloud environments):
# download data and open xarray dataset
curr_shortname = 'ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4'
ds_SSH = ea.ecco_podaac_to_xrdataset(curr_shortname,\
StartDate='2017-01',EndDate='2017-12',\
mode='download',\
download_root_dir=join(user_home_dir,'Downloads','ECCO_V4r4_PODAAC'))
created download directory /home/jovyan/Downloads/ECCO_V4r4_PODAAC/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4
DL Progress: 100%|#########################| 12/12 [00:05<00:00, 2.16it/s]
=====================================
total downloaded: 71.02 Mb
avg download speed: 12.76 Mb/s
Time spent = 5.56673002243042 seconds
We specified a root directory for the download (which also happens to be the default setting), and the data files are then placed under download_root_dir / ShortName. We can verify that the contents of the file are what we queried:
ds_SSH
<xarray.Dataset> Size: 25MB
Dimensions: (time: 12, tile: 13, j: 90, i: 90, i_g: 90, j_g: 90, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 96B 2017-01-16T12:00:00 ... 2017-12-16T0...
... ...
YC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
XG (tile, j_g, i_g) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YG (tile, j_g, i_g) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] 192B dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHIBC (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHNOIBC (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ETAN (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2017-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2017-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: a21a5c30-400c-11eb-a9e0-0cc47a3f49c3In-cloud direct access with pre-generated json files (mode = ‘s3_open_fsspec’)#
Now, if you are part of the ECCO Hackweek you are also working in a cloud environment which means that you have many more access modes open to you. Let’s try s3_open_fsspec, which opens the files from S3 (no download necessary), and uses json files with the data chunking information to open the files exceptionally fast. This means you need to provide the directory where the jsons are located, on the efs_ecco drive: /efs_ecco/mzz-jsons.
curr_shortname = 'ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4'
ds_SSH_s3 = ea.ecco_podaac_to_xrdataset(curr_shortname,\
StartDate='2017-01',EndDate='2017-12',\
mode='s3_open_fsspec',\
jsons_root_dir=join('/efs_ecco','mzz-jsons'))
ds_SSH_s3
<xarray.Dataset> Size: 25MB
Dimensions: (time: 12, tile: 13, j: 90, i: 90, nb: 4, j_g: 90, i_g: 90, nv: 2)
Coordinates: (12/13)
XC (tile, j, i) float32 421kB ...
XC_bnds (tile, j, i, nb) float32 2MB ...
XG (tile, j_g, i_g) float32 421kB ...
YC (tile, j, i) float32 421kB ...
YC_bnds (tile, j, i, nb) float32 2MB ...
YG (tile, j_g, i_g) float32 421kB ...
... ...
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 96B 2017-01-16T12:00:00 ... 2017-12-16T0...
time_bnds (time, nv) datetime64[ns] 192B ...
Dimensions without coordinates: nb, nv
Data variables:
ETAN (time, tile, j, i) float32 5MB ...
SSH (time, tile, j, i) float32 5MB ...
SSHIBC (time, tile, j, i) float32 5MB ...
SSHNOIBC (time, tile, j, i) float32 5MB ...
Attributes: (12/57)
Conventions: CF-1.8, ACDD-1.3
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 1992-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 1992-01-01T12:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
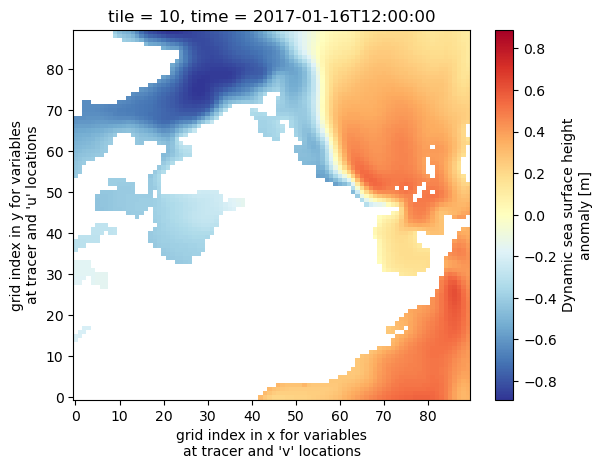
uuid: 9302811e-400c-11eb-b69e-0cc47a3f49c3Now plot the SSH for Jan 2017 in tile 10 (Python numbering convention; 11 in Fortran/MATLAB numbering convention). Here we use the “RdYlBu” colormap, one of many built-in colormaps that the matplotlib package provides, or you can create your own. The “_r” at the end reverses the direction of the colormap, so red corresponds to the maximum values.
ds_SSH_s3.SSH.isel(time=0,tile=10).plot(cmap='RdYlBu_r')
<matplotlib.collections.QuadMesh at 0x7fe336d81750>
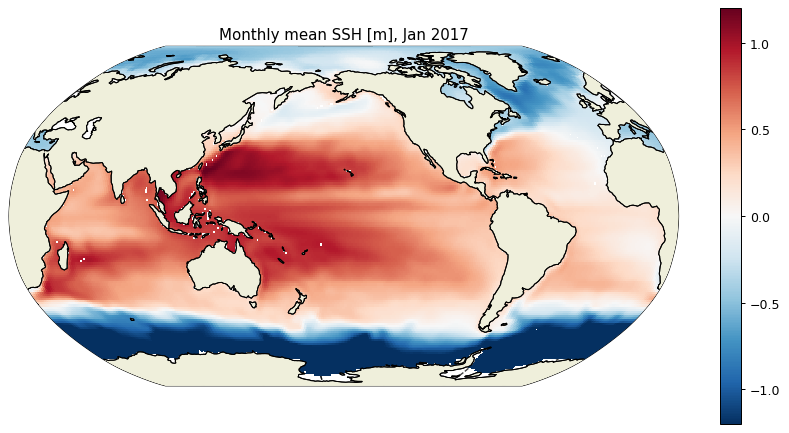
We can also use the ecco_v4_py package to plot a global map of Jan 2017 SSH, using the plot_proj_to_latlon_grid function which regrids from the native LLC grid to a lat/lon grid.
import ecco_v4_py as ecco
plt.figure(figsize=(12,6), dpi= 90)
ecco.plot_proj_to_latlon_grid(ds_SSH_s3.XC, ds_SSH_s3.YC, \
ds_SSH_s3.SSH.isel(time=0), \
user_lon_0=-160,\
projection_type='robin',\
plot_type='pcolormesh', \
cmap='RdBu_r',\
dx=1,dy=1,cmin=-1.2, cmax=1.2,show_colorbar=True)
plt.title('Monthly mean SSH [m], Jan 2017')
plt.show()
What if you don’t know the ShortName already?#
NASA Earthdata datasets are identified by ShortNames, but you might not know the ShortName of the variable or category of variables that you are seeking. One way to find the ShortName is to consult these ECCOv4r4 variable lists. But the “query” in ecco_access functions does not have to be a ShortName; it can also be a text string representing a variable name, or a word or phrase in the variable description.
For example, perhaps you are looking to open the dataset that has native grid monthly sea ice concentration in 2007. If the query is not identified as a ShortName, then a text search of the variable lists is conducted using query, grid, and time_res. Then of the identified matches, the user is asked to select one.
ds_seaice_conc = ea.ecco_podaac_to_xrdataset('ice',grid='native',time_res='monthly',\
StartDate='2007-01',EndDate='2007-12',\
mode='s3_open_fsspec',\
jsons_root_dir=join('/efs_ecco','mzz-jsons'))
ShortName Options for query "ice":
Variable Name Description (units)
Option 1: ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
SSH Dynamic sea surface height anomaly. Suitable for
comparisons with altimetry sea surface height data
products that apply the inverse barometer
correction. (m)
SSHIBC The inverted barometer correction to sea surface
height due to atmospheric pressure loading. (m)
SSHNOIBC Sea surface height anomaly without the inverted
barometer correction. Suitable for comparisons
with altimetry sea surface height data products
that do NOT apply the inverse barometer
correction. (m)
ETAN Model sea level anomaly, without corrections for
global mean density changes, inverted barometer
effect, or volume displacement due to submerged
sea-ice and snow. (m)
Option 2: ECCO_L4_STRESS_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
EXFtaux Wind stress in the model +x direction (N/m^2)
EXFtauy Wind stress in the model +y direction (N/m^2)
oceTAUX Ocean surface stress in the model +x direction,
due to wind and sea-ice (N/m^2)
oceTAUY Ocean surface stress in the model +y direction,
due to wind and sea-ice (N/m^2)
Option 3: ECCO_L4_HEAT_FLUX_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
EXFhl Open ocean air-sea latent heat flux (W/m^2)
EXFhs Open ocean air-sea sensible heat flux (W/m^2)
EXFlwdn Downward longwave radiative flux (W/m^2)
EXFswdn Downwelling shortwave radiative flux (W/m^2)
EXFqnet Open ocean net air-sea heat flux (W/m^2)
oceQnet Net heat flux into the ocean surface (W/m^2)
SIatmQnt Net upward heat flux to the atmosphere (W/m^2)
TFLUX Rate of change of ocean heat content per m^2
accounting for mass (e.g. freshwater) fluxes
(W/m^2)
EXFswnet Open ocean net shortwave radiative flux (W/m^2)
EXFlwnet Net open ocean longwave radiative flux (W/m^2)
oceQsw Net shortwave radiative flux across the ocean
surface (W/m^2)
SIaaflux Conservative ocean and sea-ice advective heat flux
adjustment, associated with temperature difference
between sea surface temperature and sea-ice,
excluding latent heat of fusion (W/m^2)
Option 4: ECCO_L4_FRESH_FLUX_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
EXFpreci Precipitation rate (m/s)
EXFevap Open ocean evaporation rate (m/s)
EXFroff River runoff (m/s)
SIsnPrcp Snow precipitation on sea-ice (kg/(m^2 s))
EXFempmr Open ocean net surface freshwater flux from
precipitation, evaporation, and runoff (m/s)
oceFWflx Net freshwater flux into the ocean (kg/(m^2 s))
SIatmFW Net freshwater flux into the open ocean, sea-ice,
and snow (kg/(m^2 s))
SFLUX Rate of change of total ocean salinity per m^2
accounting for mass fluxes (g/(m^2 s))
SIacSubl Freshwater flux to the atmosphere due to
sublimation-deposition of snow or ice (kg/(m^2 s))
SIrsSubl Residual sublimation freshwater flux (kg/(m^2 s))
SIfwThru Precipitation through sea-ice (kg/(m^2 s))
Option 5: ECCO_L4_SEA_ICE_CONC_THICKNESS_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
SIarea Sea-ice concentration (fraction between 0 and 1)
SIheff Area-averaged sea-ice thickness (m)
SIhsnow Area-averaged snow thickness (m)
sIceLoad Average sea-ice and snow mass per unit area
(kg/m^2)
Option 6: ECCO_L4_SEA_ICE_VELOCITY_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
SIuice Sea-ice velocity in the model +x direction (m/s)
SIvice Sea-ice velocity in the model +y direction (m/s)
Option 7: ECCO_L4_SEA_ICE_HORIZ_VOLUME_FLUX_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
ADVxHEFF Lateral advective flux of sea-ice thickness in the
model +x direction (m^3/s)
ADVyHEFF Lateral advective flux of sea-ice thickness in the
model +y direction (m^3/s)
DFxEHEFF Lateral diffusive flux of sea-ice thickness in the
model +x direction (m^3/s)
DFyEHEFF Lateral diffusive flux of sea-ice thickness in the
model +y direction (m^3/s)
ADVxSNOW Lateral advective flux of snow thickness in the
model +x direction (m^3/s)
ADVySNOW Lateral advective flux of snow thickness in the
model +y direction (m^3/s)
DFxESNOW Lateral diffusive flux of snow thickness in the
model +x direction (m^3/s)
DFyESNOW Lateral diffusive flux of snow thickness in the
model +y direction (m^3/s)
Option 8: ECCO_L4_SEA_ICE_SALT_PLUME_FLUX_LLC0090GRID_MONTHLY_V4R4 *native grid,monthly means*
oceSPflx Net salt flux into the ocean due to brine
rejection (g/(m^2 s))
oceSPDep Salt plume depth (m)
Using dataset with ShortName: ECCO_L4_SEA_ICE_CONC_THICKNESS_LLC0090GRID_MONTHLY_V4R4
We selected option 5, corresponding to ShortName ECCO_L4_SEA_ICE_CONC_THICKNESS_LLC0090GRID_MONTHLY_V4R4. Let’s look at the dataset contents.
ds_seaice_conc
<xarray.Dataset> Size: 25MB
Dimensions: (time: 12, tile: 13, j: 90, i: 90, nb: 4, j_g: 90, i_g: 90, nv: 2)
Coordinates: (12/13)
XC (tile, j, i) float32 421kB ...
XC_bnds (tile, j, i, nb) float32 2MB ...
XG (tile, j_g, i_g) float32 421kB ...
YC (tile, j, i) float32 421kB ...
YC_bnds (tile, j, i, nb) float32 2MB ...
YG (tile, j_g, i_g) float32 421kB ...
... ...
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 96B 2007-01-16T12:00:00 ... 2007-12-16T1...
time_bnds (time, nv) datetime64[ns] 192B ...
Dimensions without coordinates: nb, nv
Data variables:
SIarea (time, tile, j, i) float32 5MB ...
SIheff (time, tile, j, i) float32 5MB ...
SIhsnow (time, tile, j, i) float32 5MB ...
sIceLoad (time, tile, j, i) float32 5MB ...
Attributes: (12/57)
Conventions: CF-1.8, ACDD-1.3
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 1992-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 1992-01-01T12:00:00
title: ECCO Sea-Ice and Snow Concentration and Thi...
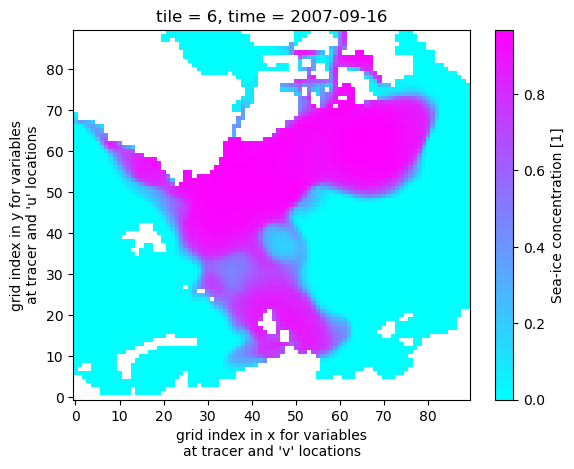
uuid: cc62f1c2-400d-11eb-9f11-0cc47a3f49c3Now plot the sea ice concentration/fraction in tile 6 (which approximately covers the Arctic Ocean), during Sep 2007 which at the time was a record minimum for Arctic sea ice.
ds_seaice_conc.SIarea.isel(time=8,tile=6).plot(cmap='cool')
<matplotlib.collections.QuadMesh at 0x7fe3363eb810>
Using the ecco_podaac_access function#
In-cloud direct access (mode = ‘s3_open’)#
The ecco_podaac_to_xrdataset function that was previously used invokes ecco_podaac_access under the hood, and ecco_podaac_access can also be called directly. This can be useful if you want to obtain a list of file objects/paths or URLs that you can then process with your own code. Let’s use this function with mode = s3_open (all s3 modes only work from an AWS cloud environment in region us-west-2).
files_dict = ea.ecco_podaac_access(curr_shortname,\
StartDate='2015-01',EndDate='2015-12',\
mode='s3_open')
{'ShortName': 'ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4', 'temporal': '2015-01-02,2015-12-31'}
Total number of matching granules: 12
files_dict[curr_shortname]
[<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-01_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-02_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-03_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-04_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-05_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-06_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-07_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-08_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-09_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-10_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-11_ECCO_V4r4_native_llc0090.nc>,
<File-like object S3FileSystem, podaac-ops-cumulus-protected/ECCO_L4_SSH_LLC0090GRID_MONTHLY_V4R4/SEA_SURFACE_HEIGHT_mon_mean_2015-12_ECCO_V4r4_native_llc0090.nc>]
The output of ecco_podaac_access is in the form of a dictionary with ShortNames as keys. In this case, the value associated with this ShortName is a list of 12 file objects. These are files on S3 (AWS’s cloud storage system) that have been opened, which is a necessary step for the files’ data to be accessed. The list of open files can be passed directly to xarray.open_mfdataset.
ds_SSH_fromlist = xr.open_mfdataset(files_dict[curr_shortname],\
compat='override',data_vars='minimal',coords='minimal',\
parallel=True)
ds_SSH_fromlist
<xarray.Dataset> Size: 25MB
Dimensions: (time: 12, tile: 13, j: 90, i: 90, i_g: 90, j_g: 90, nv: 2, nb: 4)
Coordinates: (12/13)
* i (i) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* i_g (i_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* j (j) int32 360B 0 1 2 3 4 5 6 7 8 9 ... 81 82 83 84 85 86 87 88 89
* j_g (j_g) int32 360B 0 1 2 3 4 5 6 7 8 ... 81 82 83 84 85 86 87 88 89
* tile (tile) int32 52B 0 1 2 3 4 5 6 7 8 9 10 11 12
* time (time) datetime64[ns] 96B 2015-01-16T12:00:00 ... 2015-12-16T1...
... ...
YC (tile, j, i) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
XG (tile, j_g, i_g) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
YG (tile, j_g, i_g) float32 421kB dask.array<chunksize=(13, 90, 90), meta=np.ndarray>
time_bnds (time, nv) datetime64[ns] 192B dask.array<chunksize=(1, 2), meta=np.ndarray>
XC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
YC_bnds (tile, j, i, nb) float32 2MB dask.array<chunksize=(13, 90, 90, 4), meta=np.ndarray>
Dimensions without coordinates: nv, nb
Data variables:
SSH (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHIBC (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
SSHNOIBC (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
ETAN (time, tile, j, i) float32 5MB dask.array<chunksize=(1, 13, 90, 90), meta=np.ndarray>
Attributes: (12/57)
acknowledgement: This research was carried out by the Jet Pr...
author: Ian Fenty and Ou Wang
cdm_data_type: Grid
comment: Fields provided on the curvilinear lat-lon-...
Conventions: CF-1.8, ACDD-1.3
coordinates_comment: Note: the global 'coordinates' attribute de...
... ...
time_coverage_duration: P1M
time_coverage_end: 2015-02-01T00:00:00
time_coverage_resolution: P1M
time_coverage_start: 2015-01-01T00:00:00
title: ECCO Sea Surface Height - Monthly Mean llc9...
uuid: a4955186-400c-11eb-8c14-0cc47a3f49c3
